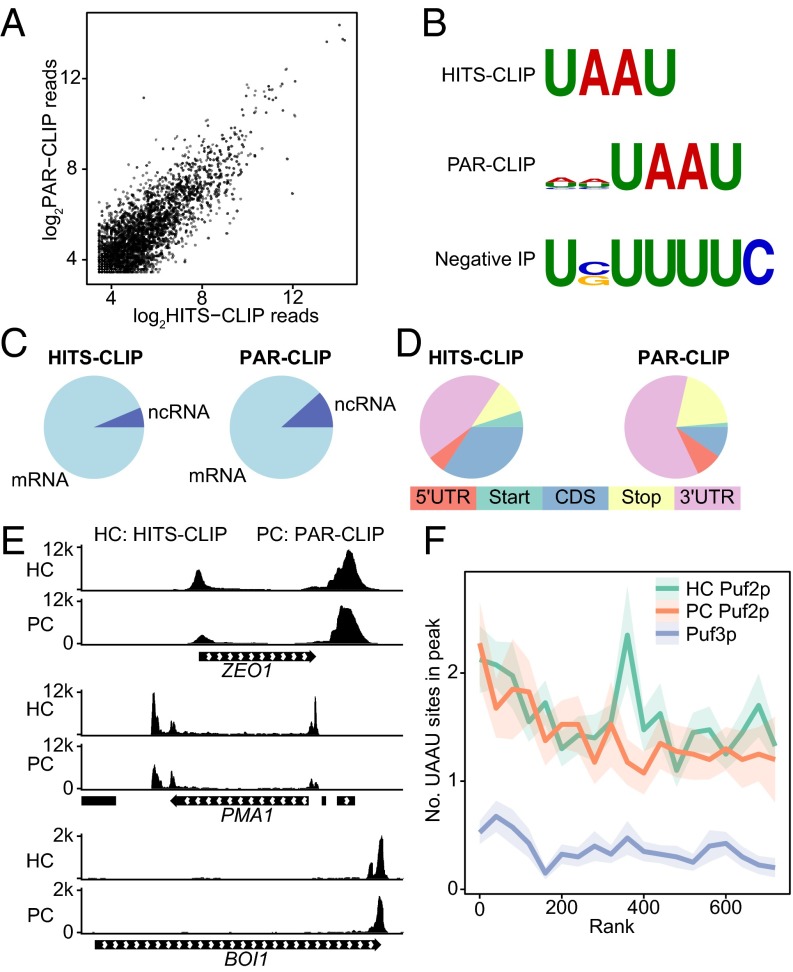
Fig. 2.
WT Puf2p binds UAAU in many targets. (A) Correlation in read depth in regions more than 10 reads depth between CLIP-seq and PAR-CLIP of Puf2p. (B) Motifs identified by DREME for untagged cells and Puf2p CLIP. “Negative IP” refers to the CLIP protocol performed on cells lacking a tagged protein. (C) Puf2p predominantly binds mRNA. (D) Puf2p binds mostly in 3′UTRs or over the stop codon. (E) Read depth per million in the two top Puf2p targets, ZEO1 and PMA1, and in an mRNA with a more common binding pattern, BOI1. Peaks occur over UAAU clusters. (F) The average number of UAAU sites in a peak as a function of gene rank. Ribbons represent SE.