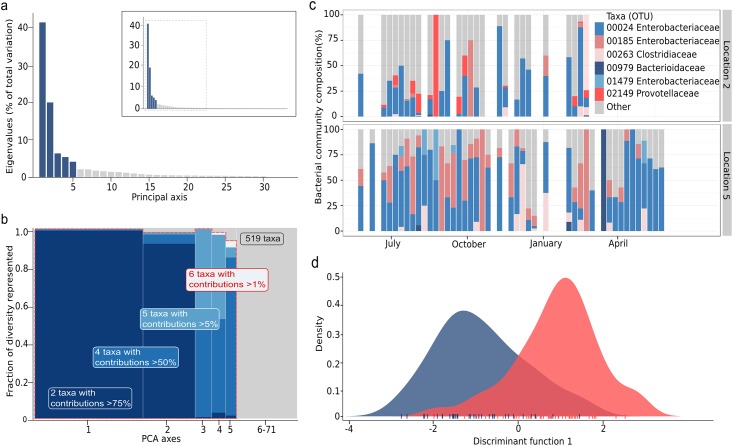
Fig 3. 16S rRNA gene surveying of gastrointestinal bacterial taxa found in stone spout and well water samples in Kathmandu, Nepal.
a) Screeplot showing the eigenvalues of the PCA of 16S rRNA gene data, with retained axes in blue corresponding to 80% of the variation in 519 identified OTUs from fecal bacterial families (Bacteroidaceae, Clostridiaceae, Enterobacteriaceae, Erysipelotrichaceae, Lachnospiraceae, Lactobacillaceae, Prevotellaceae, Ruminococcaceae and Veillonellaceae) identified in 93 water samples from location 2 (stone spout) and location 5 (sunken well). The main graph only represents the first 30 eigenvalues (full graph provided in inset). b) Diversity represented by the OTUs with large contributions to the retained PCA axes. PCA axes are represented on the x-axis, with a width proportional to the corresponding diversity (eigenvalue). The y-axis represents the amount of diversity retained by retaining only OTUs with contributions of at least 1% (6 taxa), 5% (5 taxa), 50% (4 taxa) or 75% (2 taxa), indicated by differing shades of blue. The total surface of a given color is proportional to the fraction of the total diversity represented by this set of taxa. The red dashed line identifies the set of 6 retained taxa plotted in Fig 3c, representing 76% of the total variation in the entire data. c) OTU composition of the water samples, showing the relative frequencies of the six most structuring fecal bacterial taxa identified in Fig 3b (Enterobacteriaceae (OTUs 00024, 00185 and 01479), Bacteroidaceae (OTU00979), Clostridiaceae (OTU00263), and Prevotellaceae (OTU 2149)). Their relative abundance (y-axis) is represented through time in location 2 (stone spout) and location 5 (sunken well), the two locations with the greatest estimated coliform contamination by MPN. Empty bars correspond to missing or failed samples. d) A Discriminant Analysis of Principal Components (DAPC) of the 16S rRNA gene identifying combinations of the 519 gastrointestinal OTUs differing the most between the stone spout (blue) and the sunken well (red). The OTUs exhibiting the greatest variation between these locations were OTU00263; Clostridium, OTU00185; Enterobacteriaceae, OTU00979; Bacteroides and OTU00024, Enterobacteriaceae).