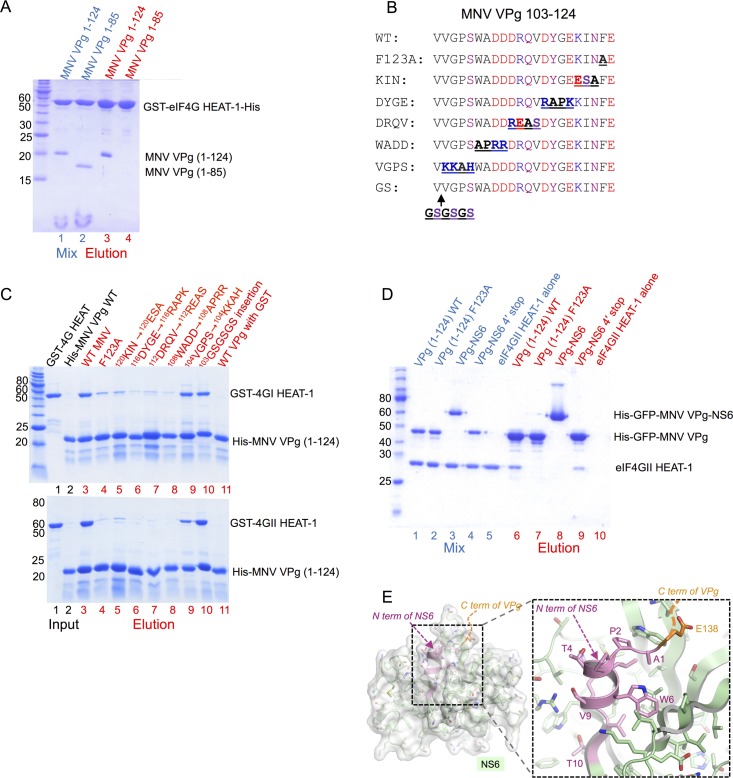
Fig 2. MNV VPg interacts with eIF4GI and eIF4GII HEAT-1 domains via the C-terminal residues 104–124.
(A) SDS PAGE analysis of cobalt affinity pull-down assay using a GST-eIF4GI HEAT-1 construct with a C-terminal His-tag as bait and either untagged MNV VPg(1–124) or MNV VPg(1–85) as prey. Protein mixtures are shown in lanes 1 and 2 (blue labels); bound proteins eluted with 250 mM imidazole are in lanes 3 and 4 (red labels). (B) Sequence alignment showing the location of amino acid substitutions introduced into the C terminus of His-tagged MNV VPg. (C) SDS PAGE analysis of cobalt affinity assays using His-tagged MNV mutants (panel B) as bait and either GST-eIF4GI-HEAT-1 (top panel) or GST-eIF4GII-HEAT-1 as prey. Lanes 1–2: input proteins; lanes 3–11: eluted proteins. (D) SDS PAGE analysis of cobalt affinity pull-down assay using either His-tagged GFP-VPg(1–124) wild-type, GFP-VPg(1–124) F123A, a His-tagged GFP-VPg-NS6 fusion (containing the inactivating C139A mutation of the protease active site Cys (NS6 numbering)) or a His-tagged GFP-VPg-NS6-4´ fusion (containing just the first 4 amino acids of NS6) as bait, and untagged eIF4GII HEAT-1 as prey. Lanes 1–5: protein mixes; lanes 6–10: proteins eluted with 250 mM imidazole. (E) Structure of the VPg-NS6 junction showing that the N-terminus of NS6 is tightly folded into the body of the protease. Structure shown is the crystal structure of VPg-NS6 from human Norwalk virus, which is very similar to the structure of MNV NS6 [40, 41]. Note that due to disorder in the crystal, only the C terminus of VPg is visible [41].