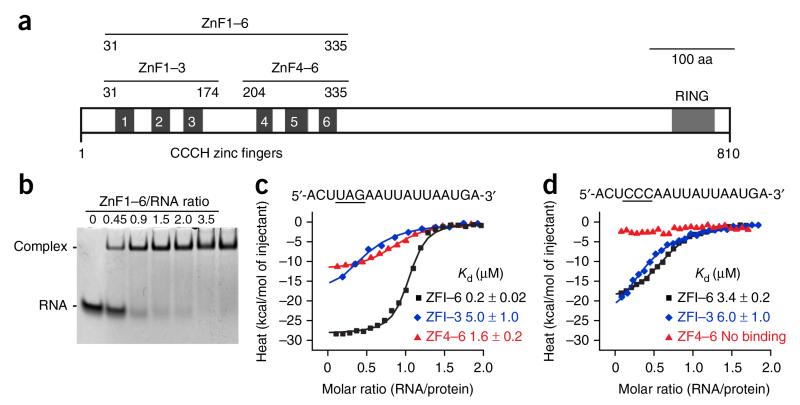
Figure 1.
Domain architecture of Unkempt and RNA affinity of its CCCH ZnFs. (a) Schematic of mouse Unkempt protein depicting all six predicted CCCH ZnFs and the RING domain. Also shown are domain boundaries of Unkempt constructs ZnF1–6, ZnF1–3 and ZnF4–6 used in this study. Aa, amino acids. (b) RNA EMSA demonstrating near-equimolar binding of recombinant ZnF1–6 to the 18-mer URE located in the HSPA8 mRNA29. The synthetic RNA was used at 40 μM. The uncropped image of the gel is shown in Supplementary Data Set 1. (c,d) ITC binding curves of complex formation between the indicated wild-type (c) or UAG-mutated (d) HSPA8 RNA and ZnF1–6, ZnF1–3 and ZnF4–6 of Unkempt. Solid lines represent nonlinear least-squares fit to the measured titration data, with binding enthalpy (kcal/mol), association constant and number of binding sites per monomer as variables. The calculated values for Kd (mean ± range of two independent technical replicates) are indicated.