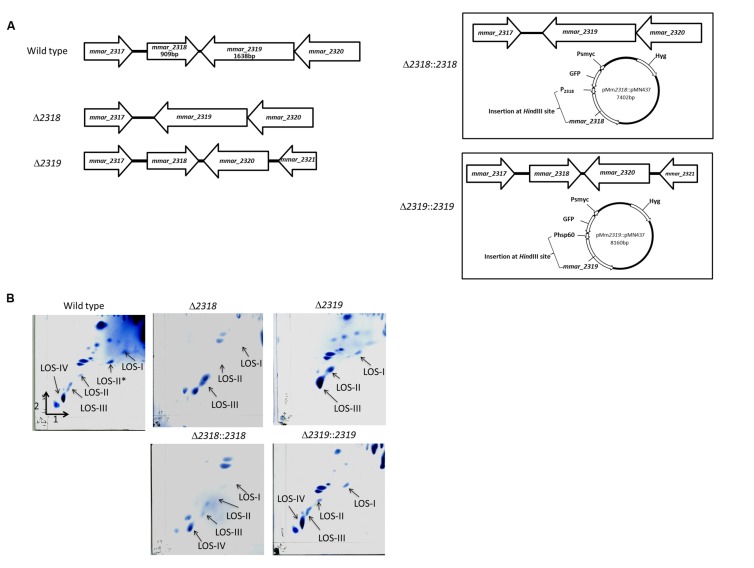
FIGURE 3.
The 2D-TLC profile of the polar lipid of M. marinum wild-type, deletion mutants and complementation strains. (A) The gene alignment of mmar_2318 and mmar_2319 deletion mutants and complementation strains. The Δ2318 and Δ2319 deletion mutants were created by unmarked deletion of mmar_2318 and mmar_2319, respectively. pMm2318::pMN437 and pMm2319::pMN437 plasmids were used to create the epichromosomal complementation strains Δ2318::2318 and Δ2319::2319, respectively. (B) The 2D-TLC profile of the polar lipid of M. marinum. Using 2D-TLC, extracted M. marinum polar lipids were separated by chloroform/methanol/water (60:30:6, v/v/v) in the first direction and by chloroform/acetic acid/methanol/water (40:25:3:6, v/v/v/v) in the second direction. The plates were charred with ceric ammonium molybdate. Accumulation of LOS-III and deficiency of LOS-IV were observed in the Δ2318 and Δ2319 mutants. The 2D-TLC lipid composition profiles of the Δ2318::2318 and Δ2319::2319 complementation strains were restored to that of the wild-type strain.