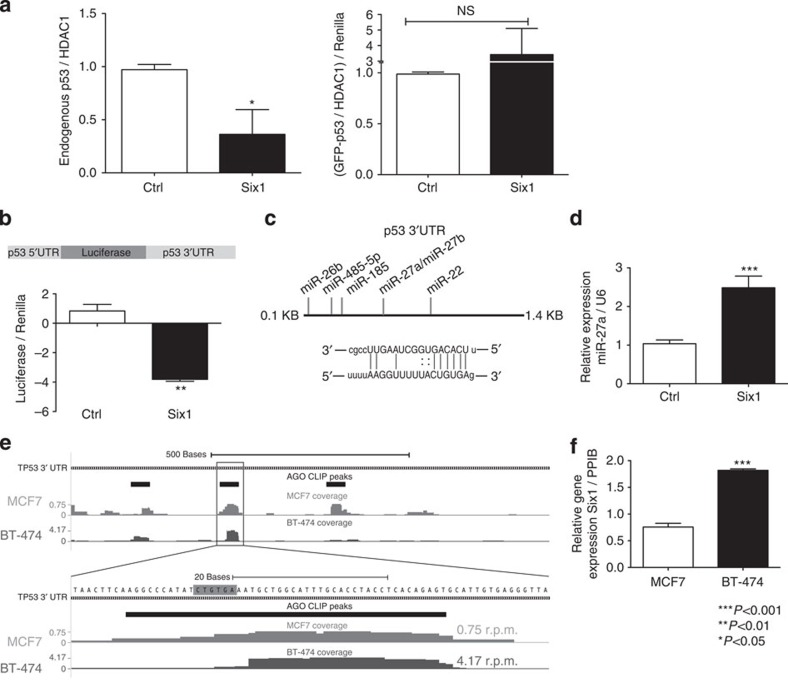
Figure 5. Six1 downregulates p53 via a microRNA-mediated mechanism.
(a) MCF7-Ctrl and Six1 cells were co-transfected with GFP–p53 and a constitutively active renilla-luciferase construct. After 1 h of etoposide treatment (10 μM), cell pellets were divided to obtain nuclear lysates (NLs) and perform luciferase assays. Endogenous p53 (left) and GFP–p53 bands (right) were quantified and normalized to endogenous HDAC1 levels. Transfected GFP–p53 was additionally normalized to the renilla luciferase values to control for transfection efficiency. Combination of 2 independent experiments, each run with experimental duplicates. (b) MCF7-Six1 or Ctrl cells were transfected with a luciferase construct containing 145 base pairs of the p53 5′-UTR and the entire p53 3′-UTR (top), as well as with a constitutively active renilla luciferase construct. T-test on mean±s.d of biological triplicates for a representative experiment (>3 experiments). (c) Top: a diagram of the microRNAs predicted to bind the p53 3′-UTR31 that were upregulated by Six1 via a microRNA microarray18. Bottom: the predicted binding of miR-27a to the p53 3′-UTR31. (d) qRT–PCR for mature miR-27a expression in MCF7-Six1 and Ctrl cells. T-test on mean±s.d. of biological triplicates for a representative experiment (three experiments). (e) The 3′-UTR of p53 showing representative HITS-CLIP coverage in BT-474 cells and MCF7 cells. Statistically significant Ago footprints (peaks) are shown above the coverage. Coverage scale (left) is in reads per million mapped reads (r.p.m.). The sequence complementary to the miR-27a-3p seed site is highlighted in grey. (f) qRT–PCR for Six1 expression in BT-474 cells as compared with MCF7 cells. T-test on mean±s.e.m. of biological triplicates for a representative experiment (two experiments).