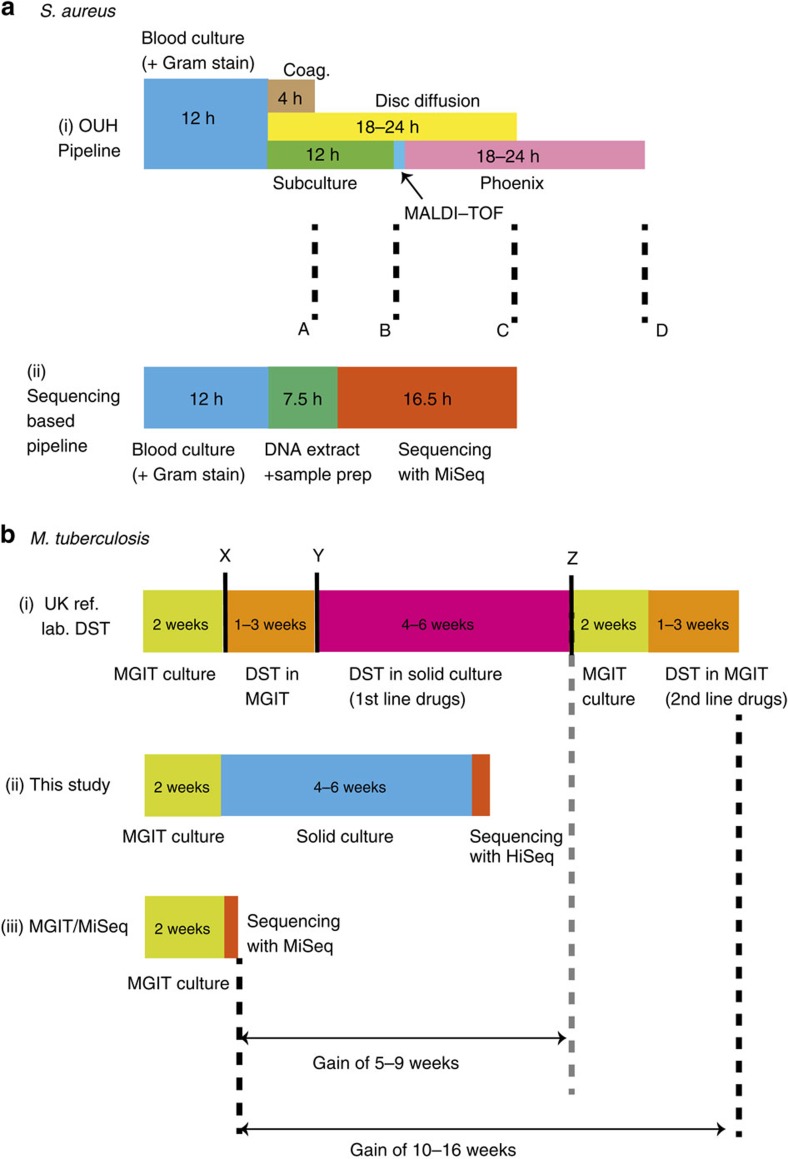
Figure 7. Timelines for sequencing-based analysis and culture-based DST.
The timelines are shown for (a) S. aureus and (b) M. tuberculosis. In (a) both culture-based (a,i) and sequencing-based (a,ii) options involve 12 h of blood culture. After this, the culture-based approach (at Oxford University Hospitals clinical laboratory) follows with a direct coagulase test (Coag.) that provides a presumptive species identification at 4 h (marked ‘A'). Concurrently, blood culture is subcultured to blood agar, and MALDI-TOF confirms the species at 12 h (‘B'). A disc diffusion test for five antimicrobials (including methicillin) is performed directly from a positive blood culture providing first-line susceptibility information 18–24 h later (‘C'), assuming an acceptable inoculum. Finally, post-subculture samples are undergo extended susceptibility testing by automated broth microdilution (brandname ‘Phoenix'), giving final results after another 18–24 h (‘D'). For the sequencing-based workflow (a,ii), the DNA extraction plus sample preparation takes 7.5 h because samples are from blood culture, not colony isolates. With the Illumina MiSeq v3 reagents, a 16.5 h run is possible (giving paired 75 bp reads, adequate for this purpose), giving full susceptibility results at the same time as direct disc tests provide results for five drugs. (b) The culture-based process (b,i; in a typical UK reference laboratory) starts with two weeks of mycobacterial growth indicator tube (MGIT) culture, followed by a species identification test (‘X'). If the species belongs to the MTBC, then DST is run in MGIT, and at decision point ‘Y', if the sample tests susceptible to all first-line drugs, no further testing is done. MGIT DST is repeated for pyrazinamide if the first test revealed resistance to this drug. If there is resistance to any other drug, then solid culture DST is performed. If these tests show there is resistance to rifampicin then another round of MGIT culture followed by MGIT DST is done for second-line drugs. For sequencing-based approaches we show timelines for the present study (b,ii) and a potential alternative (b,iii), which would reduce time-to-results to just over 2 weeks.