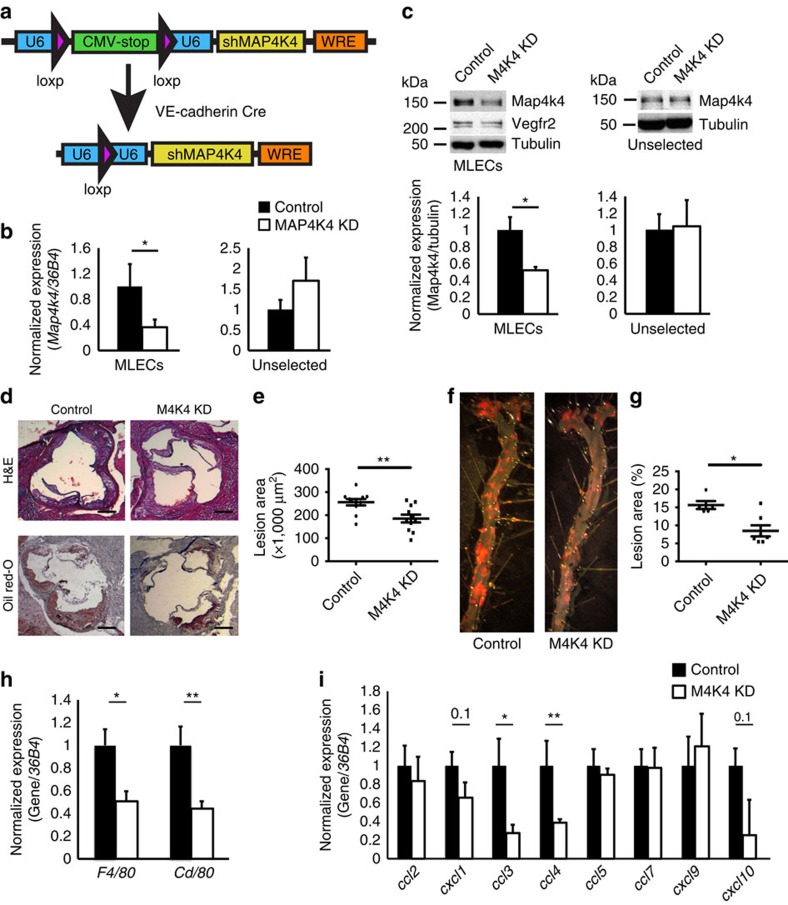
Figure 3. Reduced atherosclerosis in M4K4 KD mice.
(a) Schematic of the transgenic construct used to generate MAP4K4 KD animals. The U6 promoter becomes reconstituted after cre-mediated recombination to drive tissue-specific shRNA expression. (b,c) Primary lung endothelial cells (MLECs) were derived from control and MAP4K4 KD animals. (b) Messenger RNA (mRNA) was extracted and quantitative RT–PCR was performed for Map4k4 in immune-selected or unselected cells. The data represent the mean±s.e.m. as normalized to 36b4 expression (*P<0.05, N=7). (c) Primary endothelial or unselected cell lysates were immunoblotted for Map4k4 and Vegfr2. Data represent the mean±s.e.m. as normalized to tubulin expression (*P<0.05, N=3–6). (d–g) MAP4K4 KD mice were crossed with Apoe−/− mice and fed a WD for 16 weeks. (d) Aortic root sections of control and MAP4K4 KD animals stained with haematoxylin and eosin (H&E) (top) and Oil Red-O (bottom). Scale bar, 250 μm. (e) Quantification of aortic root lesion area. Data represent the mean±s.e.m. (**P<0.005, N=9,11). (f) Oil Red-O-stained en face aortic preparations from control and MAP4K4 KD animals. (g) Quantification of Oil red-O-stained area. Data represent the mean±s.e.m. (*P<0.05, N=5–7). (h–i) mRNA was prepared from whole aortas, and qPCR was performed. (h) Macrophage markers F4/80 and Cd68. (i) Chemokines Ccl2, Cxcl1, Ccl3, Ccl4, Ccl5, Ccl7, Cxcl9 and Cxcl10. Data represent the mean±s.e.m. as normalized to 36b4 (*P<0.05, **P<0.005, N=9–10).