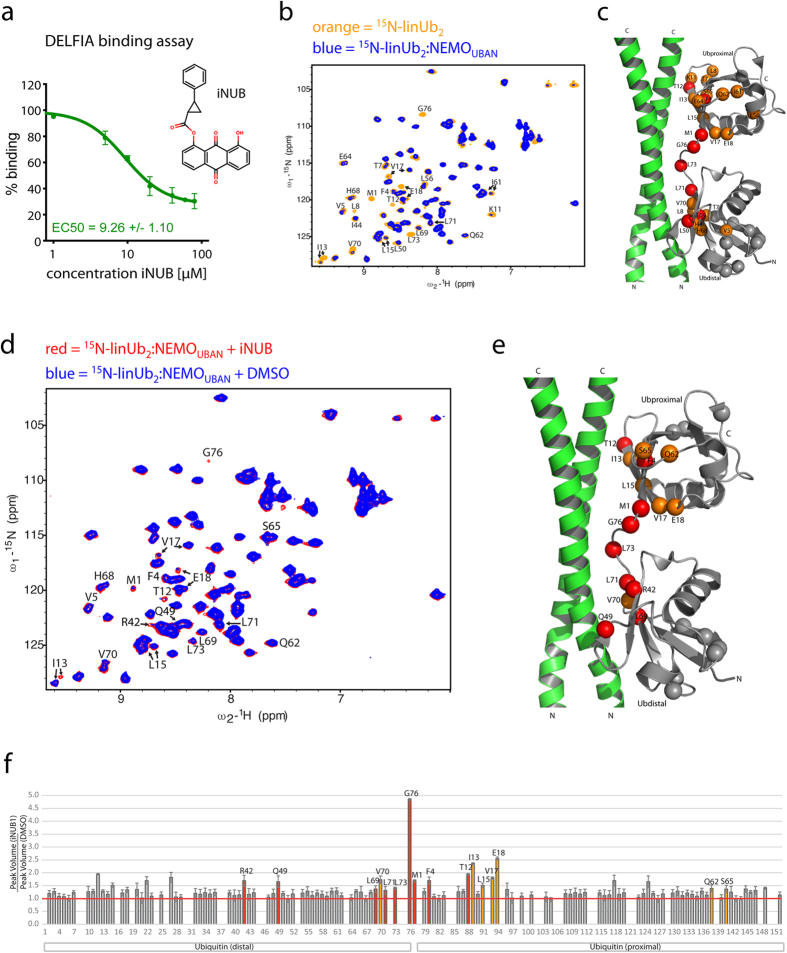
Figure 3. The small molecule iNUB inhibits binding of NEMOUBAN to linUb2.
(a) DELFIA assay detection of StrepTagII-NEMOUBAN (242–350) binding to His-tagged linUb2 with or without iNUB. Increasing iNUB concentrations disrupted binding of NEMO to linUb2 binding with an EC50 of 9.26 μM. (b) NMR analysis of the NEMOUBAN-linUb2 interaction. 1H, 15N HSQC spectra of 100 μM 15N-labeled linUb2 in absence (orange) and in presence of unlabeled NEMOUBAN C347S (258–350) (blue) at 1:1 stoichiometry. (c) Backbone amide resonances of Ub2 resonances showing strongest changes upon NEMOUBAN addition are annotated and indicated as spheres on the NEMO crystal structure (PDB 2ZVO). Red spheres indicate residues with unambiguous chemical shift assignments, while orange color shows amides, which could not be unambiguously assigned to the proximal or distal Ub moieties or showing signal overlap. Gray spheres are the amides of the corresponding residues in the other Ub module. (d) 1H, 15N HSQC spectra of 100 μM 15N-labeled Ub2 bound to the NEMOUBAN (1:1) in the presence of DMSO (blue) or 290 μM iNUB (red). NMR signals of linUb2 residues that showed reduced signal intensity upon binding to NEMOUBAN are partially restored upon treatment with iNUB. (e) Mapping of Ub residues that show strongest effects upon iNUB addition onto the NEMOUBAN structure as in (d). (f) Ratio of amide peak volume upon iNUB versus DMSO addition for the 15N-labeled Ub2 complexed with unlabeled NEMO (1:1 stoichiometry).