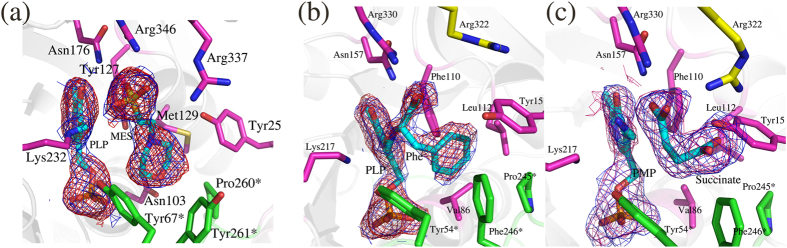
Figure 3. Ligand recognition by mHspAT and mArAT.
(a) The electron density (ED) maps (2|Fo|–|Fc| (blue) and |Fo|–|Fc| (red) at 1σ and 2.5σ contour levels respectively) for the holo form shows MES bound near PLP. ED maps (2|Fo|–|Fc| (blue) and |Fo|–|Fc| (red) at 1σ and 2.5σ contour levels, respectively are shown for PLP-Phe (b) and PMP and succinate molecules (c) bound in the active site of mArAT complexes. The two distinct substrates are recognized by mArAT using the “arginine switch” mechanism, in which Arg322 (yellow) moves away from the active site to accommodate the neutral phenyl ring of Phe.