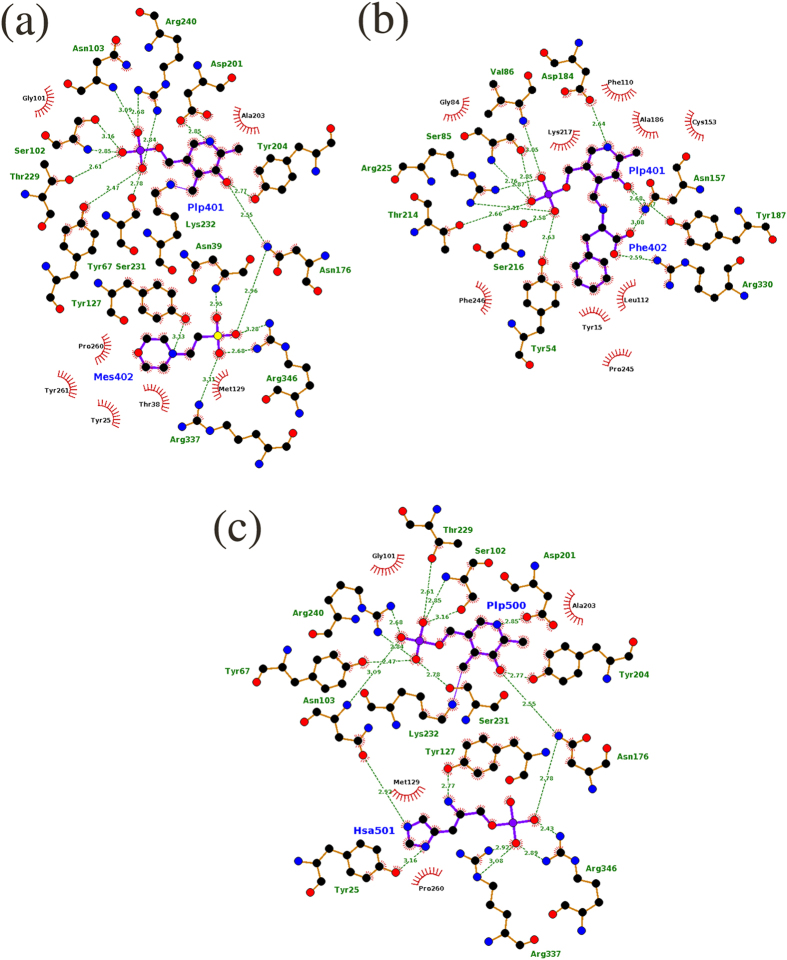
Figure 6.
Schematic representations of the atomic interactions between (a) mHspAT and PLP-MES, (b) mArAT and PLP-Phe and (c) mHspAT and PLP-Hsp (Hsa). In both enzymes, PLP makes stronger binding than the substrate. Residues which are involved in H-bond interaction (shown in green dotted lines with the corresponding donor-acceptor distance) are shown in ball and stick model, whereas those that are involved in van der Waals interactions with the ligands are shown in spikes. In panel (a), Tyr67, Pro260, and Tyr261 protrude from the adjacent molecule (chain B; PDB ID: 4R8D) of the dimer. Try54, Pro245 and Phe246, in panel (b), belong to the other molecule (chain D; PDB ID: 4R2N) of the dimer). In panel (c), Tyr67 and Pro260 protrude from the adjacent molecule.