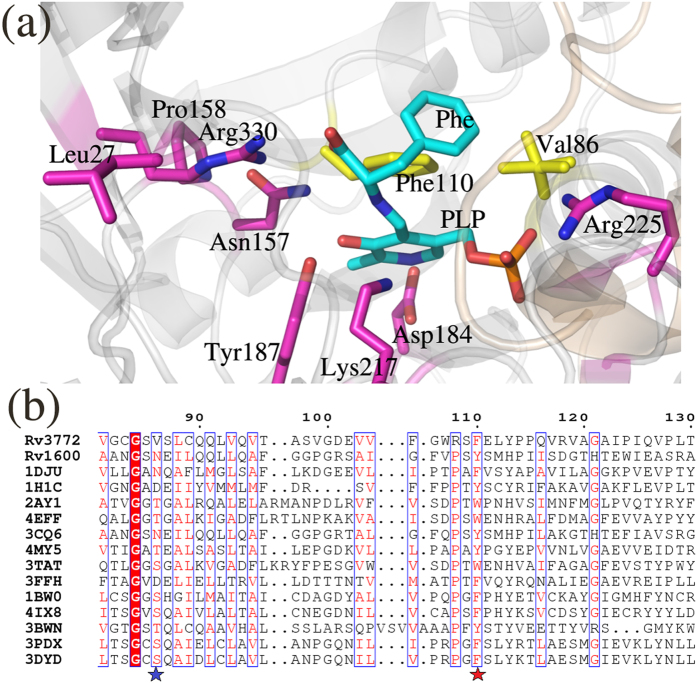
Figure 8. Structural and sequence alignment of selected aminotransferases of subfamily Iβ with mHspAT and mArAT.
The source of each sequence and PDB ID are: Pyrococcus horikoshii ArAT (1DJU)6, T. maritima HspAT (1H1C)8, Paracoccus denitrificans ArAT (2AY1)18, Burkholderia pseudomallei ArAT (4EFF), C. glutamicum HspAT (3CQ6)9, Streptococcus mutans ArAT (4MY5), E. coli TyrAT (3TAT)21, Listeria innocua HspAT (3FFH), Trypanosoma cruzi TyrAT (1BW0)20, Leishmania infantum TyrAT (4IX8)22, Arabidopsis thaliana TrpAT (3BWN)23, Mouse TyrAT (3PDX)24 and Human TyrAT (3DYD). (a) A close up view of the active site of PLP- mArAT is shown with the strictly conserved residues in magenta colour, whereas the residues which are critical for substrate binding and are partially conserved (0.7 consensus) are shown in yellow. (b) A segment of alignment shows that the equivalent positions of Val86 of mArAT is largely occupied by polar residues (marked with a blue star) and equivalent positions of Phe110 of mArAT is occupied by only aromatic amino acids (marked with a red star). The residues that are strictly conserved among the homologous sequences are highlighted in filled-red boxes. Whereas the residue positions showing 70% consensus are highlighted in blue-frame boxes with the similar residues shown in red letters.