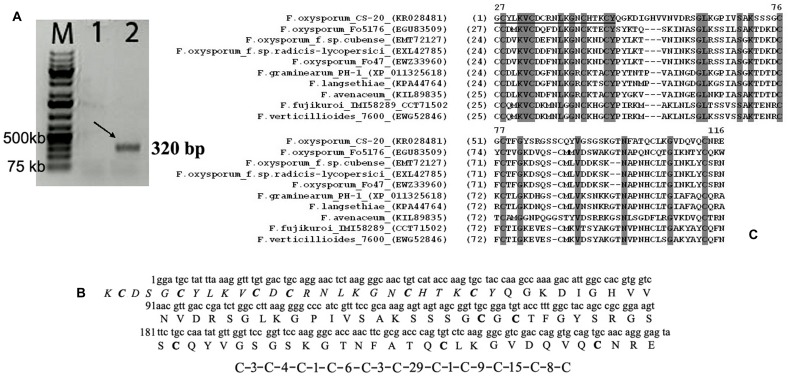
FIGURE 10.
A representative result of electrophoretic analysis of PCR-products amplified by two-step 3′ RACE (A); nucleotide and deduced amino acid sequences of the elicitor protein from F. oxysporum CS-20 – CS20EP (B); as well as the alignment of predicted CS20EP amino acid sequence with related sequences deposited in GenBank (C). (A) The arrow indicates the target PCR-product of an estimated size (320 bp) that was amplified after the second 3′RACE round with AdLo-CS20F1 primer pair at different annealing temperatures. Lane 1, 60°C; line 2, 52°C; M, GeneRulerTM 1 kb DNA ladder plus. (B) The predicted amino acid sequence of the CS-20 elicitor protein (CS20EP) is shown below the nucleotide sequence (GenBank accession number KR028481). The fifty-five amino acid residues corresponding to those determined by N-terminal sequencing of the protein are marked in italic. Cysteine residues in the amino acid sequence are marked in bold. The cysteine motif is presented under the sequence. (C) Identical amino acid residues in sequences are highlighted by dark-gray boxes. The CS20EP N-terminal sequence determined by Edman’s degradation is underlined.