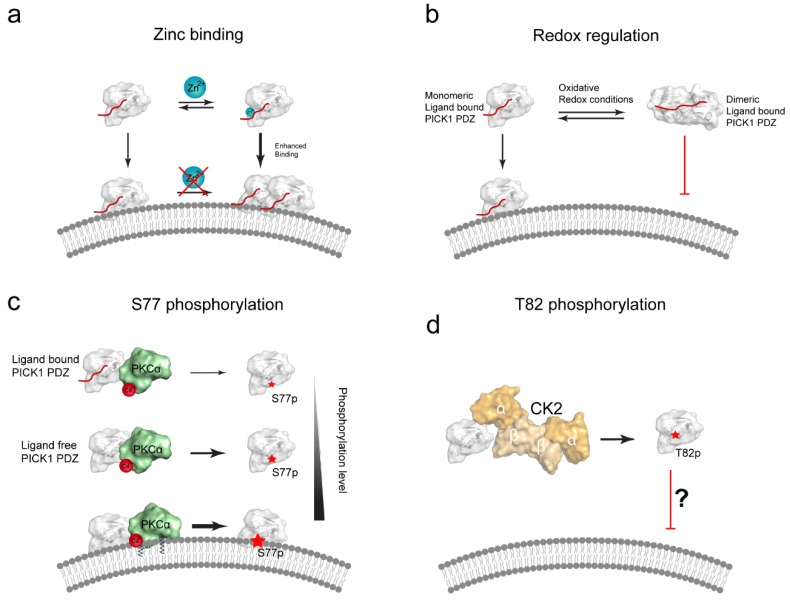
Figure 4.
Proposed regulatory mechanisms for the PICK1 PDZ membrane binding. (a) Zn2+-binding has been suggested to positively regulate the membrane binding of the isolated, ligand bound PICK1 PDZ domain. This interaction was shown to involve the Cys44 and Cys46 in the CPC loop. Remarkably, Zn2+-binding could be completely abolished when the PDZ domain was already bound to liposomes, suggesting that this mechanism does not require/allow Zn2+ to be inserted into the membrane [78]; (b) Proposed mechanism for redox regulation of the PICK1 PDZ domain. PICK1 forms reversible dimeric complexes under oxidative conditions, via interdomain cystines involving Cys44 and Cys46. In contrast to the monomeric ligand bound (ligand shown in red) PDZ domain, the dimeric ligand bound PDZ domain complex is not capable of binding liposomes (Dimeric PICK1-PDZ PDB:3HPK) [73]; (c) Ser77, positioned in the cationic patch, is a PKCα (shown in green, PDB: 3GPE [79]) substrate [77]. Phosphorylation of Ser77 in full length PICK1 was decreased in presence of a PDZ domain ligand, compared to a ligand free PDZ domain. Conversely, the phosphorylation level was significantly enhanced in the presence of liposomes. A decrease in clustering in COS7 cells suggested that the S77D phospho-mimetic could weaken the PDZ:lipid interaction in cells; (d) T82, positioned at the edge of the binding groove, is a putative phosphorylation site for CK2 (shown in shades of orange, PDB: 1JWH [80]). T82E likewise displays decrease cellular PICK1:GluA2 co-clusters, in a mechanism possible alleviating subcellular localization by reduced PDZ domain lipid binding capacity.