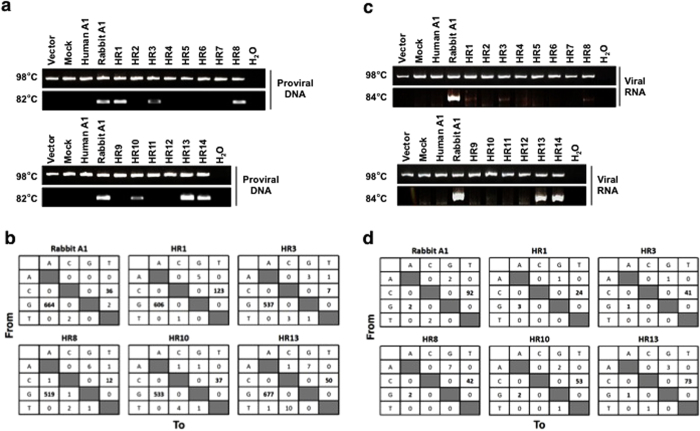
Figure 6. Selective amplification of edited HIV-1 proviral genomes and genomic viral RNA.
(a) The isolated DNA was amplified using primer sets specific for the tat/env region of NL4-3. The nested PCR products denatured at 98 °C were compared with those denatured at 82 °C, after separation with 2% agarose gel electrophoresis. After staining with ethidium bromide, the gel was photographed. (b) Mutation matrices of G-to-A and C-to-T mutations in the tat/env gene in the proviral DNA of Vif-proficient virus amplified with PCR after denaturation at 82 °C. The boxes show the differences between the nucleotide sequence of the tat/env gene and the observed sequence in the presence of rabbit A1 or the indicated chimeric A1 proteins (n = 12, 6,384 bp). (c) Isolated viral RNA was amplified using primer sets specific for the tat/env region of NL4-3. The nested PCR products denatured at 98 °C were compared with those denatured at 84 °C, after separation with 2% agarose gel electrophoresis. After staining with ethidium bromide, the gel was photographed. (d) Mutation matrices of G-to-A and C-to-U mutations in the genomic RNA of Vif-proficient virus. PCR was performed with denaturation at 84 °C. The boxes show the differences between the nucleotide sequence of the tat/env gene and the observed sequence in the absence of A1 or the presence of human, rabbit, or chimeric A1 protein (n = 12, 6,384 bp).