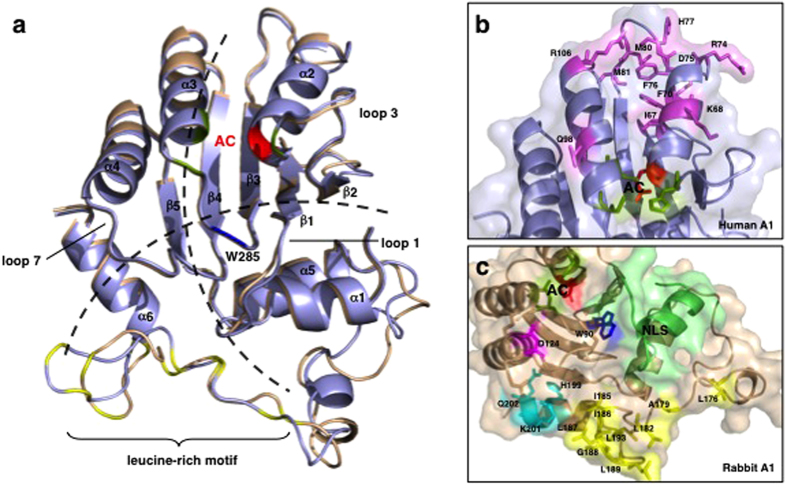
Figure 7. Comparison of the structures of the human and rabbit A1 proteins.
(a) Homology models of human A1 (light blue) and rabbit A1 (wheat) show structural conservation relative to A3G-CTD (Fig. S1). The ribbon schematics show the core five hydrophobic β-sheets (β1–β5) surrounded by six α-helices (α1–α6). The loop structures (loops 1, 3, and 7), predicted to form between adjacent α-helices and β-sheets, have been implicated in the interaction between the protein and its nucleic acid substrates. The two proposed models of nucleic acid binding to the proteins are represented by dotted lines. In the Zn2+-coordinating active core (AC), cysteine (green) and histidine (red) residues are indicated. W285 (blue) was reported to be involved in the enzyme’s deamination activity in A3G-CTD and is conserved in human and rabbit A1s. The C-terminal regions of both A1s contain a leucine-rich motif (leucine residues colored yellow). (b) Residues in human A1 that differ from those in rabbit A1 in the substituted region of chimera HR14 are shown as sticks (violet). (c) In rabbit A1, residue D124 (magenta), implicated in the local dinucleotide preference of A3G, is conserved but the residues surrounding it differ from those in human A1. The NLS located in the N-terminal region of the protein is shown in lime green. The leucine-rich motif (Leu and aliphatic residues; yellow) located in the C-terminal region is followed by residues that differ from those in human A1 (H199, K201, and Q202; cyan). Homology models were built with SWISS-MODEL and the images were generated with PyMol.