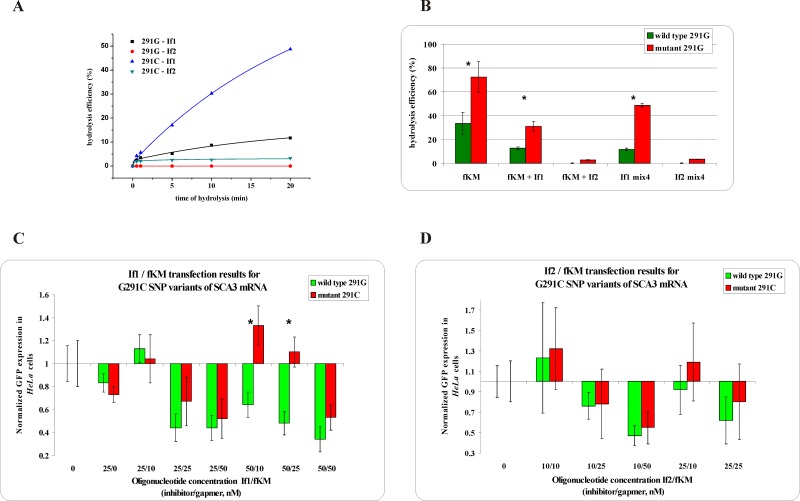
Fig 3. The RNase H assay results for C692G SNP within APP mRNA.
(A) kinetics of the WT and Mut RNAs cleavage in the presence of the gapmer (bKM) and the shorter (Ib1) or longer (Ib2) inhibitor, where black squares indicate degradation of WT RNA in the presence of the Ib1/bKM/Mut RNA mixture, red dots—degradation of WT RNA in the presence of the Ib2/bKM/ Mut RNA mixture, blue triangles—degradation of Mut RNA in the presence of the Ib1/bKM/WT RNA mixture and green triangles—degradation of Mut RNA in the presence of the Ib2/bKM/WT RNA mixture, (B) stability of the WT RNA (green bar) and Mut RNA (red bar) in the presence of: gapmer bKM only (first pair of bars from the left), gapmer bKM and short inhibitor Ib1 (second) or gapmer bKM and longer inhibitor Ib2 (third), and in the WT/Mut RNA/bKM/Ib1 mixture (fourth) and WT/Mut RNA/bKM/Ib2 mixture (fifth); statistically significant differences between the mean hydrolysis efficiency of the RNA variants (P<0.05) are marked with asterisk, (C,D) Results of HeLa cells cotransfection with WT/Mut C692G-pEGFP constructs and different amounts of inhibitor and gapmer antisense oligonucleotides. qPCR results of tandem approach with (C) shorter inhibitor Ib1, (D) Longer inhibitor Ib2. Statistically significant differences between the mean of the RNA variants expression (P<0.05) are marked with asterisk.