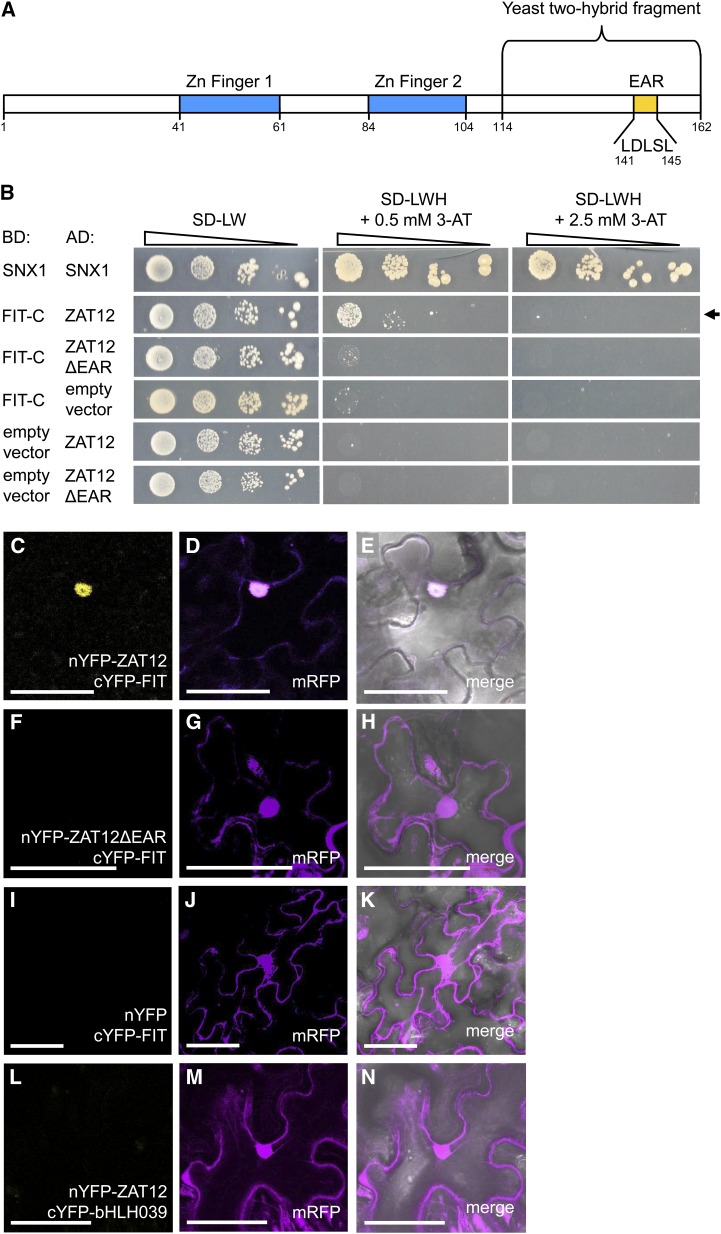
Figure 1.
ZAT12 interaction with FIT and requirement of the EAR motif. A, Schematic representation of the ZAT12 amino acid sequence. The regions encoding the two Zn fingers, the EAR motif with five amino acids removed in the deletion studies, and the ZAT12 fragment of 48 amino acids identified in the yeast two-hybrid screen are indicated. B, Targeted yeast two-hybrid interaction assay for FIT-C and ZAT12 protein interaction. Ten-fold dilutions series (A600 of 1−1–10−4) of the yeast AH109 strain, harboring a combination of a DNA-binding domain (BD) vector and an activation domain (AD) vector, were spotted on synthetic defined (SD) selection medium without Leu and Trp (-LW; selection for positive cotransformants) or without Leu, Trp, and His (-LWH) with added 0.5 or 2.5 mm 3-amino-1,2,4-triazole (3-AT; selection for interaction) and incubated for 5 to 8 d at 30°C. The combination of BD-SNX1 and AD-SNX1 was used as a positive control. BD-FIT-C and AD-ZAT12 or AD-ZAT12ΔEAR were assayed for interaction. Their combinations with the respective empty vectors were used as negative controls. The positive interaction between BD-FIT-C and AD-ZAT12 is indicated by the arrow. C to N, rBiFC analysis on N. benthamiana leaf epidermis cells transiently transformed with nYFP-ZAT12 and cYFP-FIT (C–E), nYFP-ZAT12ΔEAR and cYFP-FIT (F–H), nYFP and cYFP-FIT (I–K), and nYFP-ZAT12 and cYFP-bHLH039 (L–N). Positive YFP signals indicative of a protein-protein interaction were detected in the nuclei of the nYFP-ZAT12 and cYFP-FIT combination but not in the combinations nYFP-ZAT12ΔEAR and cYFP-FIT or nYFP-ZAT12 and cYFP-bHLH039, or in the empty vector control nYFP and cYFP-FIT. The mRFP signal was present in the nuclei and in the cytoplasm of all transformed cells. C, F, I, and L show YFP fluorescence; D, G, J, and M show mRFP fluorescence; and E, H, K, and N show merged YFP, mRFP, and bright-field images. Bars = 50 µm.