Abstract
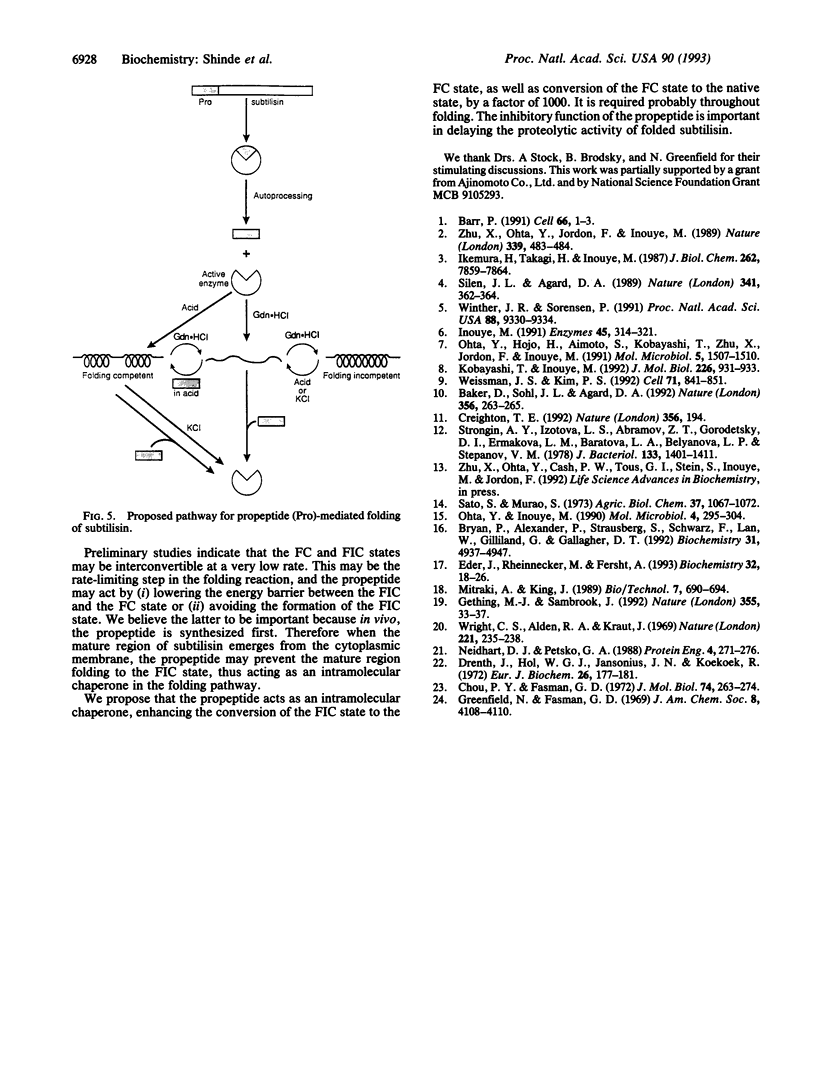
The N-terminal propeptide of subtilisin, a serine protease, functions as an intramolecular chaperone which is crucial for proper folding of the active enzyme. This nascent N-terminal propeptide is removed after completion of the folding process. Here we present a possible pathway by which intramolecular chaperones mediate protein folding. Using circular dichroism to analyze acid-denatured subtilisin we have identified a folding-competent state which can refold to an active conformation in the absence of the propeptide. Earlier work had shown that guanidine hydrochloride-denatured subtilisin was in a state incapable of folding in absence of its propeptide. Comparison of the folding-incompetent and folding-competent states indicates that refolding is facilitated by the presence of residual structure present only in the folding-competent state. The analysis further indicates that the propeptide is essential for inducing this state. Therefore the folding-competent state may lie on--or be in rapid equilibrium with an intermediate on--the folding pathway of subtilisin. In the absence of the propeptide, formation of such a state--and hence refolding--is extremely slow.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker D., Sohl J. L., Agard D. A. A protein-folding reaction under kinetic control. Nature. 1992 Mar 19;356(6366):263–265. doi: 10.1038/356263a0. [DOI] [PubMed] [Google Scholar]
- Barr P. J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991 Jul 12;66(1):1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Bryan P., Alexander P., Strausberg S., Schwarz F., Lan W., Gilliland G., Gallagher D. T. Energetics of folding subtilisin BPN'. Biochemistry. 1992 Jun 2;31(21):4937–4945. doi: 10.1021/bi00136a003. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Structural and functional role of leucine residues in proteins. J Mol Biol. 1973 Mar 5;74(3):263–281. doi: 10.1016/0022-2836(73)90372-0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Protein folding. Up the kinetic pathway. Nature. 1992 Mar 19;356(6366):194–195. doi: 10.1038/356194a0. [DOI] [PubMed] [Google Scholar]
- Drenth J., Hol W. G., Jansonius J. N., Koekoek R. Subtilisin Novo. The three-dimensional structure and its comparison with subtilisin BPN'. Eur J Biochem. 1972 Mar 27;26(2):177–181. doi: 10.1111/j.1432-1033.1972.tb01754.x. [DOI] [PubMed] [Google Scholar]
- Eder J., Rheinnecker M., Fersht A. R. Folding of subtilisin BPN': characterization of a folding intermediate. Biochemistry. 1993 Jan 12;32(1):18–26. doi: 10.1021/bi00052a004. [DOI] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969 Oct;8(10):4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Ikemura H., Takagi H., Inouye M. Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli. J Biol Chem. 1987 Jun 5;262(16):7859–7864. [PubMed] [Google Scholar]
- Inouye M. Intramolecular chaperone: the role of the pro-peptide in protein folding. Enzyme. 1991;45(5-6):314–321. doi: 10.1159/000468904. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Inouye M. Functional analysis of the intramolecular chaperone. Mutational hot spots in the subtilisin pro-peptide and a second-site suppressor mutation within the subtilisin molecule. J Mol Biol. 1992 Aug 20;226(4):931–933. doi: 10.1016/0022-2836(92)91042-n. [DOI] [PubMed] [Google Scholar]
- Neidhart D. J., Petsko G. A. The refined crystal structure of subtilisin Carlsberg at 2.5 A resolution. Protein Eng. 1988 Oct;2(4):271–276. doi: 10.1093/protein/2.4.271. [DOI] [PubMed] [Google Scholar]
- Ohta Y., Hojo H., Aimoto S., Kobayashi T., Zhu X., Jordan F., Inouye M. Pro-peptide as an intramolecular chaperone: renaturation of denatured subtilisin E with a synthetic pro-peptide [corrected]. Mol Microbiol. 1991 Jun;5(6):1507–1510. doi: 10.1111/j.1365-2958.1991.tb00797.x. [DOI] [PubMed] [Google Scholar]
- Ohta Y., Inouye M. Pro-subtilisin E: purification and characterization of its autoprocessing to active subtilisin E in vitro. Mol Microbiol. 1990 Feb;4(2):295–304. doi: 10.1111/j.1365-2958.1990.tb00596.x. [DOI] [PubMed] [Google Scholar]
- Strongin A. Y., Izotova L. S., Abramov Z. T., Gorodetsky D. I., Ermakova L. M., Baratova L. A., Belyanova L. P., Stepanov V. M. Intracellular serine protease of Bacillus subtilis: sequence homology with extracellular subtilisins. J Bacteriol. 1978 Mar;133(3):1401–1411. doi: 10.1128/jb.133.3.1401-1411.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman J. S., Kim P. S. The pro region of BPTI facilitates folding. Cell. 1992 Nov 27;71(5):841–851. doi: 10.1016/0092-8674(92)90559-u. [DOI] [PubMed] [Google Scholar]
- Winther J. R., Sørensen P. Propeptide of carboxypeptidase Y provides a chaperone-like function as well as inhibition of the enzymatic activity. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9330–9334. doi: 10.1073/pnas.88.20.9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. S., Alden R. A., Kraut J. Structure of subtilisin BPN' at 2.5 angström resolution. Nature. 1969 Jan 18;221(5177):235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]
- Zhu X. L., Ohta Y., Jordan F., Inouye M. Pro-sequence of subtilisin can guide the refolding of denatured subtilisin in an intermolecular process. Nature. 1989 Jun 8;339(6224):483–484. doi: 10.1038/339483a0. [DOI] [PubMed] [Google Scholar]