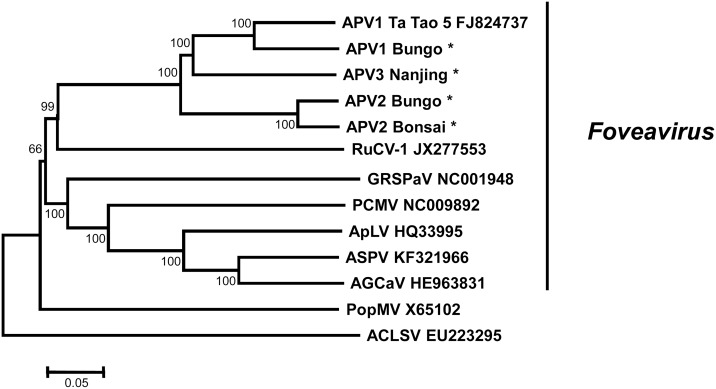
Fig 2. Unrooted phylogenetic tree reconstructed with the complete genome sequence of representative members of the family Betaflexiviridae.
Tree was constructed by the neighbor-joining method and the statistical significance of branches was evaluated by bootstrap analysis (1,000 replicates). Only bootstrap values above 65% are indicated. The scale bar represents 5% nucleotide divergence. The abbreviations followed by the accession numbers are: APV1, 2 or 3, Asian prunus virus 1, 2 or 3, respectively; RuCV-1, Rubus canadensis virus 1; GRSPaV, Grapevine rupestris stem pitting associated virus; PCMV, Peach chlorotic mottle virus; ApLV, Apricot latent virus; ASPV, Apple stem pitting virus; AGCaV, Apple green crinkle associated virus; PopMV, Poplar mosaic virus; ACLSV, Apple chlorotic leaf spot virus. The sequences of APV1, 2, and 3 determined in this work are identified by an asterisk *.