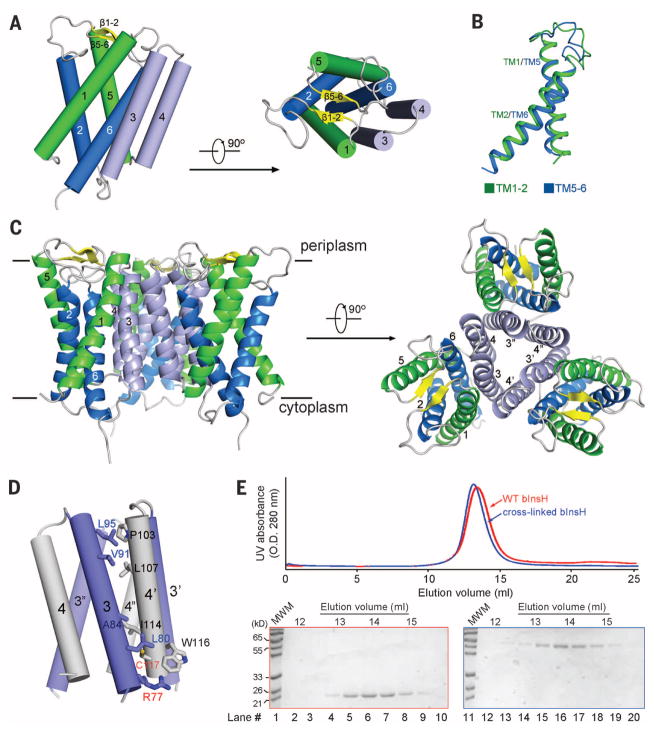
Fig. 1. The crystal structure of MvINS, a mycobacterial homolog of mammalian Insig proteins.
(A) The overall structure of an MvINS monomer. Two perpendicular views are shown. It reveals a novel fold that may be shared by all Insig proteins. (B) TM1/TM2 can be superimposed to TM5/TM6. (C) Three adjacent MvINS molecules form a homotrimer in the crystal. The side and periplasmic views of the overall trimer are shown. (D) The trimeric interface is mediated exclusively by hydrophobic residues on TM3 and TM4. Arg77 on TM3 was mutated to Cys for disulfide-bond formation with Cys117 on TM4 from the adjacent protomer. Both residues are labeled red. (E) MvINS is a trimer in solution. Wild-type and cross-linked MvINS-R77C were subjected to size exclusion chromatography. The peak fractions were applied to SDS–polyacrylamide gel electrophoresis followed by Coomassie blue staining. All structure figures were prepared with PyMol (37).