FIGURE 3.
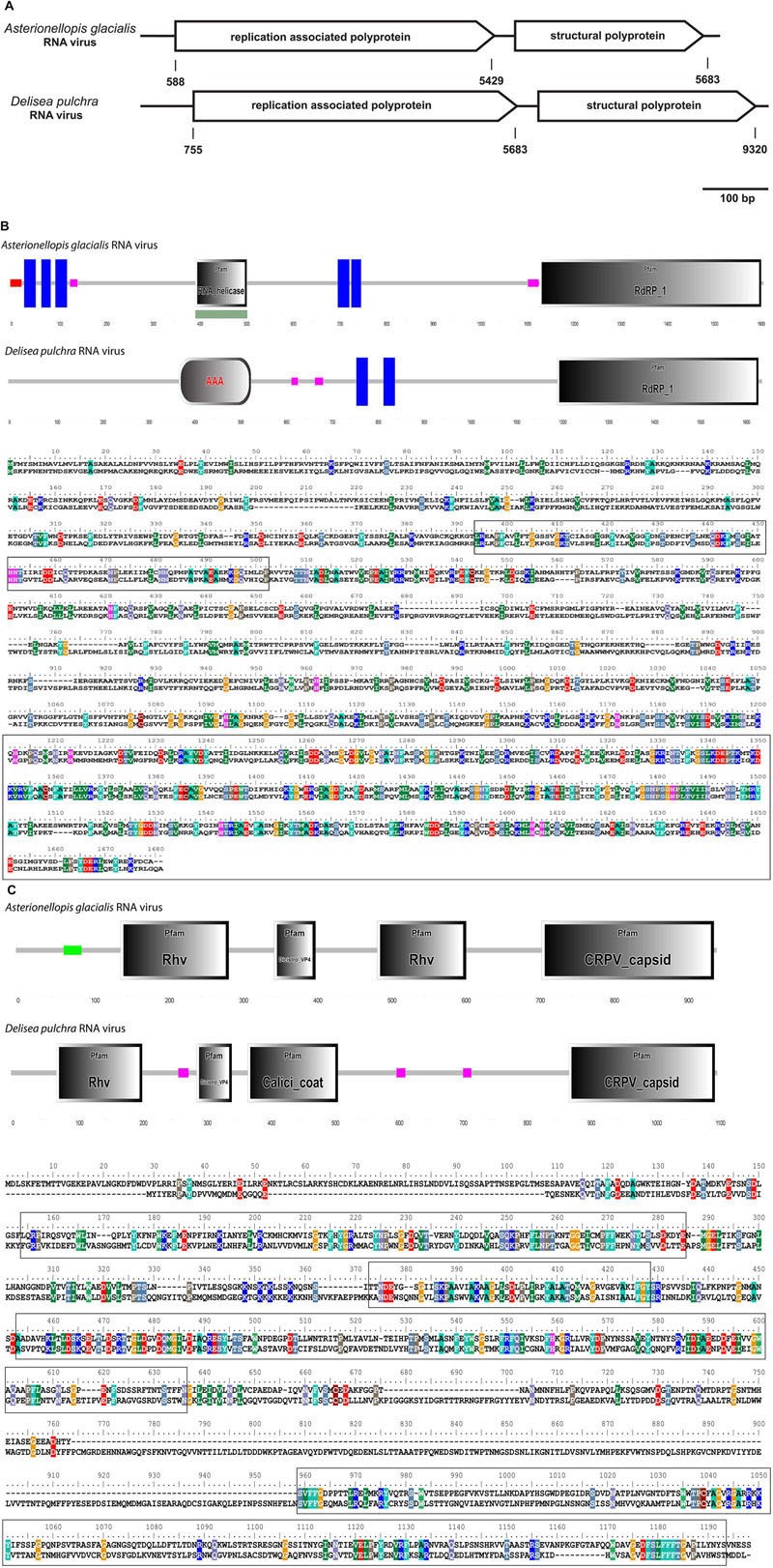
(A–C) Comparison between the Asterionellopis glacialis (“Agla”) RNA virus and the D. pulchra RNA virus. (A) The genomes of both viruses encode two polyproteins, one replication associated and one structural polyprotein. (B) Protein domain analysis using SMART (Simple Modular Architecture Research Tool) and sequence alignment of the replication associated polyprotein shows a similar structural organization consisting of RNA helicase or replication associated protein AAA, transmembrane proteins (blue boxes) and RdRP. Low complex regions are shown in pink. (C) Protein domain analysis and sequence alignment of the structural polyprotein shows four structural protein domains Rhv, DicistroVP4, Calici coat, and CRPV capsid. The Calici coat domain overlaps with the second Rhv domain of the Agla RNA virus. Sequences of conserved protein domains are highlighted in boxes.
