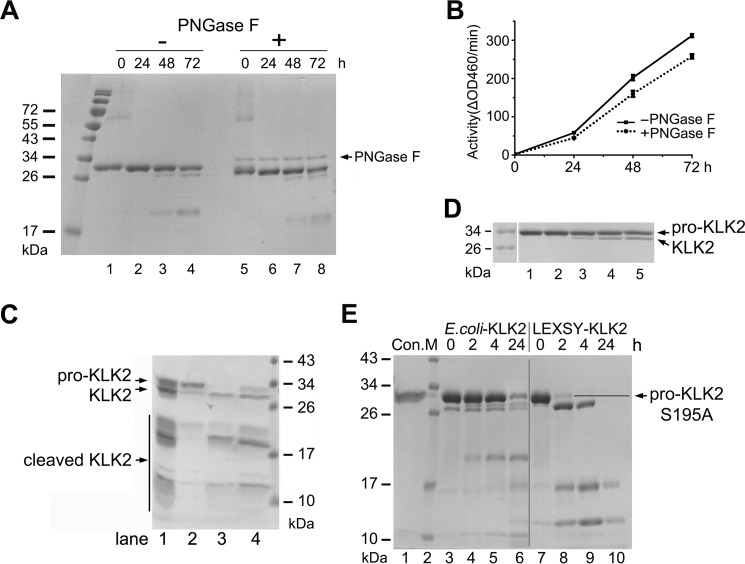
FIGURE 2.
Autoactivation and activation of pro-KLK2. A, time-dependent autoactivation of both glycosylated (lanes 1–4) and enzymatically deglycosylated pro-KLK2 (lanes 5–8) is monitored on an SDS-PAGE. Note the same shift of about 1 kDa as in Fig. 1B. B, activity assay of autoactivated KLK2 with the small peptide substrate H-PFR-AMC by glycosylated (black line) and enzymatically deglycosylated KLK2 (dotted line) at varying times (0, 24, 48, and 72 h), matching those in A. C, concentration-dependent autoactivation of pro-KLK2-S195A. The initial pro-KLK2 samples at concentrations of 250 (lane 1), 600 (lane 2), 800 (lane 3), and 900 (lane 4) μg/ml were kept at room temperature for 1 day and subsequently at 4 °C for 4 days, respectively. D, activation of pro-KLK2. Shown is incubation of 1 μg of pro-KLK2-S195A from LEXSY cells (lane 1) with 1% KLK2e from E. coli (lane 2), 1% glyco-KLK2 from LEXSY cells (lane 3), or 0.05% (lane 4) or 0.1% enterokinase (lane 5) overnight (14 h) at room temperature. E, pro-KLK2-S195A (lane 1) was digested by 20% KLK2e (lanes 3–6) or 20% glyco-KLK2 (lanes 7–10) for a time-dependent course. Samples were taken at 0, 2, 4, and 24 h, as indicated above the lanes, and then checked by Coomassie Blue-stained SDS-PAGE. The vertical gray line was inserted to indicate samples with different treatment. KLK2e shows a different cleavage pattern (e.g. by generating a 20-kDa fragment, which originates from the clipping at Arg-70↓His-71).