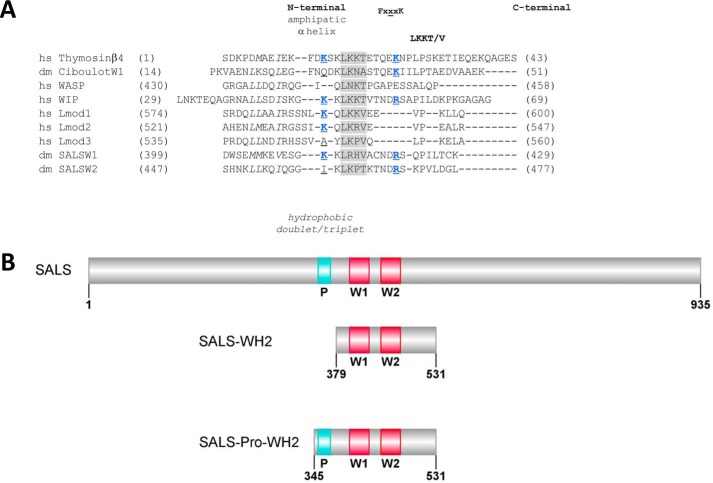
FIGURE 1.
Comparative sequence analysis of the WH2 domains of SALS and domain organization of the constructs investigated in this study. A, comparative sequence alignment of the WH2 domains of SALS. The hydrophobic amino acid doublet/triplet in the N-terminal region is shown by italics; the consensus LKK(T/V) motif is highlighted in gray; amino acids shown previously to form salt bridges with actin are shown in blue; and the central amino acid of the FXXXK region is underlined. hs, Homo sapiens; dm, D. melanogaster. Lmod, leiomodin; WASP, Wiskott-Aldrich syndrome protein. Sequence IDs are as follows: Thymosinβ4 (NP_066932.1); Ciboulot (NP_525065.1); WASP (NP_000368.1); WIP (O43312); Lmod1 (NP_036266); Lmod2 (NP_997046.1); Lmod3 (NP_938012), and SALS (NP_001163588.1). B, SALS constructs used in this study. P, proline-rich sequence region; W1 and W2, WH2 domains. The figure was made by Illustrator for BioSequence software (12).