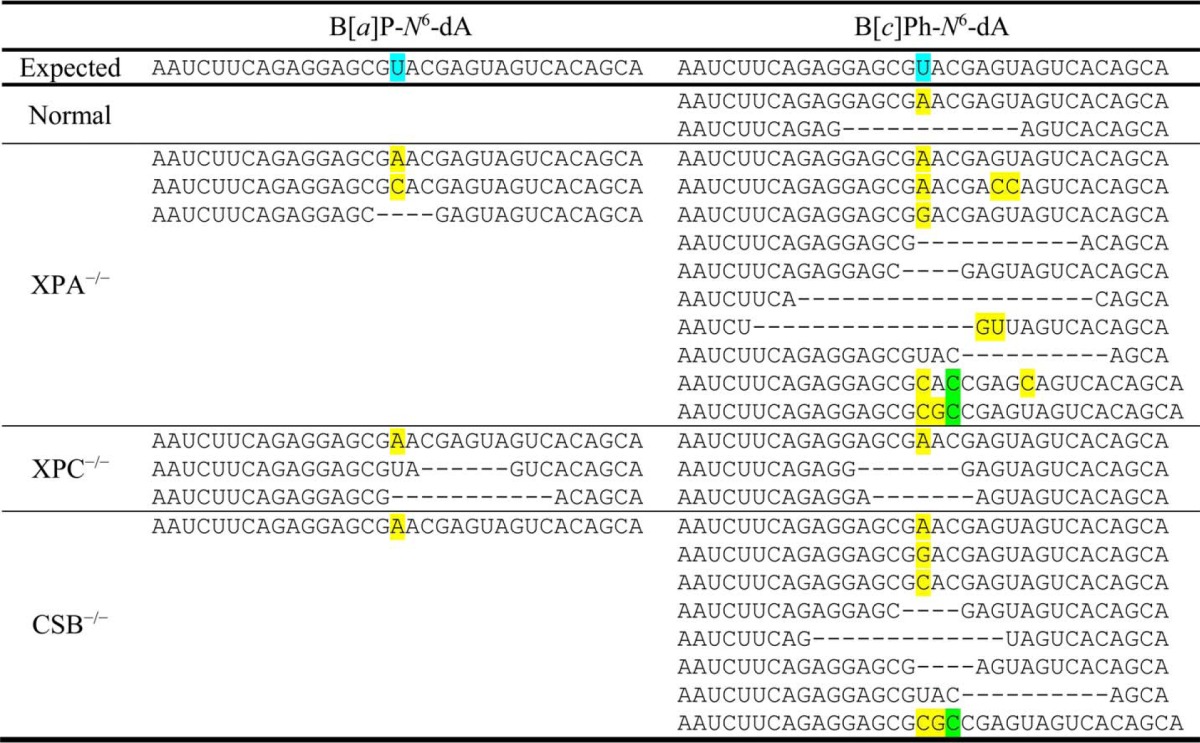
TABLE 3.
Representative base misincorporations, deletions and insertions observed in mRNA following transcription past B[a]P-N6-dA or B[c]Ph-N6-dA
The mRNA data shown were generated by amplifying cDNA with barcoded primers containing Illumina HiSeq adapters, and sequencing the resulting amplicons on a HiSeq 2500 System (Illumina, San Diego, CA). Analysis of the resultant reads was accomplished with custom Perl scripts. The mRNA shown represent accumulated, abnormal transcripts that were observed at a frequency ≥10 times the background misincorporation rate of 0.1% in each sample. Only the portions of the full-length transcripts in the vicinity of the DNA adduct are shown. Bases highlighted in yellow indicate sites of misincorporation; bases highlighted in green indicate insertions; and a dash indicates the site of deletions.(Additional experiments are needed to understand how the pattern of abnormal transcripts changes quantitatively over time.) Normal cells are proficient in both global NER and TCR; XPA−/− cells are deficient in both global NER and TCR; XPC−/− cells are deficient in global NER but proficient in TCR; and CSB−/− cells are proficient in global NER but deficient in TCR. The expected mRNA sequence is shown, with the U marked in blue indicating the complementary position within the DNA template's transcription unit that was modified with either B[a]P-N6-dA or B[c]Ph-N6-dA. Note that in normal, DNA repair-proficient fibroblasts, no abnormal transcripts were observed; in all other fibroblasts, normal mRNA was found along with the abnormal transcripts.