Figure 1.
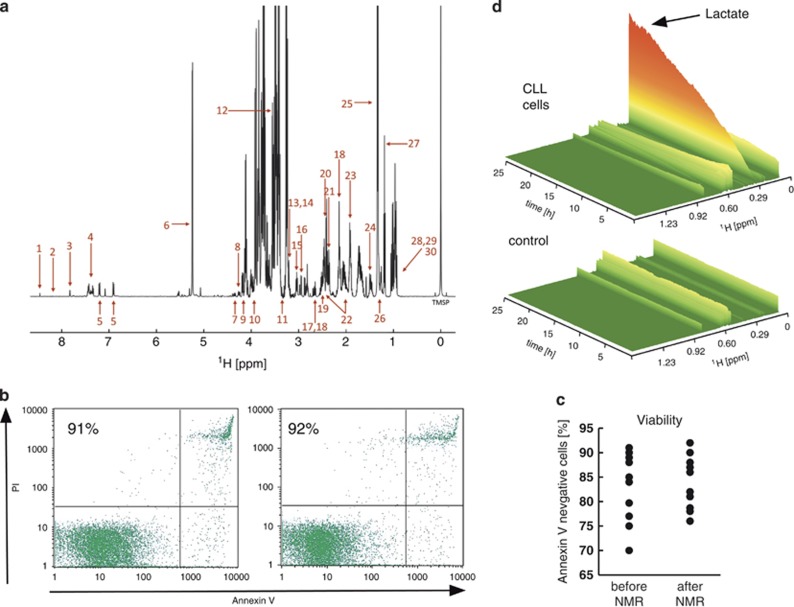
CLL cells survive NMR analyzes and display marked metabolic activity. Primary-CLL mononuclear cells were monitored for 24 h in the NMR at 37°C. (a) Representative one-dimensional-1H NMR spectrum of CLL cells in 0.1% agarose in serum-free bicarbonate buffered RPMI 1640 growth medium. Metabolites assigned: 1-formate, 2-hypoxanthine*, 3-histidine, 4-phenylalanine, 5-tyrosine, 6-glucose, 7-trans-4-hydroxyl-L-proline, 8-uridine, 9-pyroglutamate, 10-serine, 11-myo-inositol, 12-glycine, 13-phosphocholine, 14-choline, 15-lysine, 16-asparagine, 17-aspartate, 18-methionine, 19-glutamine, 20-succinate, 21-pyruvate, 22-glutamate, 23-arginine, 24-alanine, 25-lactate, 26-3-hydroxybutyrate, 27-ethanol, 28-valine, 29-isoleucine and 30-leucine. *hypoxanthine was detectable after a few hours of the time course. Data shown is representative of >25 primary CLL samples. Viability of CLL cells was assessed pre- and post-NMR analysis by (b) monitoring cell morphology of Jenner–Giemsa stained cell cytopsins, and (c) Annexin V/propidium iodide (PI) staining and flow cytometry. Viable cells are identifiable as Annexin V/PI negative (lower left quadrant). Percentage viable cells is indicated in the scatter plot. Data shown is representative of >25 samples. (c) Viability data for 10 primary CLL samples. (d) Representative three-dimensional view of an NMR time course experiment. The control sample contained RPMI medium with ITS+ and 0.1% low melting point agarose (no cells), whereas the second panel contained additionally 5 × 107 CLL cells/ml. Metabolite intensity is highlighted by a color gradient and height. The tallest visible orange peak corresponds to lactate. Data shown are representative of three CLL samples.