Figure 1.
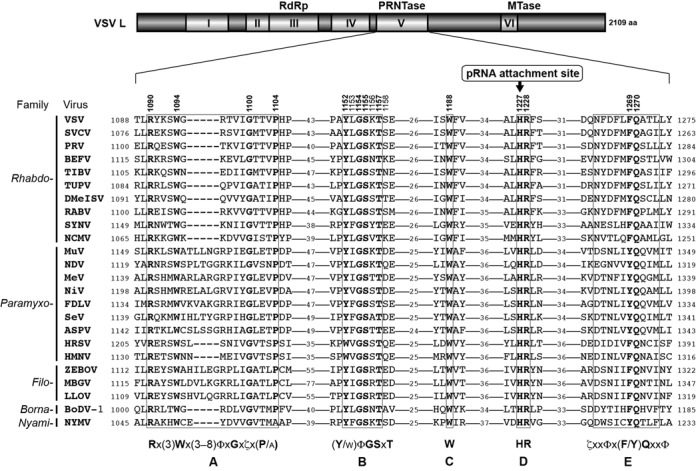
Conserved amino acid sequence motifs of the PRNTase domain in NNS RNA viral L proteins. A schematic structure of the L protein of VSV (Vesiculovirus) is shown with six conserved amino acid sequence blocks (I–VI) (28). RdRp, PRNTase and MTase indicate RNA-dependent RNA polymerase, GDP polyribonucleotidyltransferase and methyltransferase domains, respectively. The amino acid sequences of block V (PRNTase domain) of representative NNS RNA viral L proteins were aligned using the PSI-Coffee program (50) (see Supplementary Figure S1), and local sequences are shown with the numbers of intervening residues. Five consensus sequences are designated as motifs A–E (x, Φ and ζ indicate any, hydrophobic and hydrophilic amino acids, respectively). The numbers above the VSV L sequence show the positions of mutated amino acid residues in the VSV L protein. H1227 in motif D has been identified as a covalent pRNA attachment site (16). Virus names (virus genera) are as follows: SVCV, spring viremia of carp virus (Sprivivirus); PRV, perch rhabdovirus (Perhabdovirus); BEFV, bovine ephemeral fever virus (Ephemerovirus); TIBV, Tibrogargan virus (Tibrovirus); TUPV, Tupaia rhabdovirus (Tupavirus), DMelSV, Drosophila melanogaster sigma virus (Sigmavirus); RABV, rabies virus (Lyssavirus); SYNV, sonchus yellow net virus (Nucleorhabdovirus); NCMV, northern cereal mosaic virus (Cytorhabdovirus); MuV, mumps virus (Rubulavirus); NDV, Newcastle disease virus (Avulavirus); MeV, measles virus (Morbillivirus); NiV, Nipah virus (Henipavirus); FDLV, Fer-de-Lance virus (Ferlavirus); SeV, Sendai virus (Respirovirus); ASPV, Atlantic salmon paramyxovirus (Aquaparamyxovirus); HRSV, human respiratory syncytial virus (Pneumovirus); HMPV, human metapneumovirus (Metapneumovirus); ZEBOV, Zaire ebolavirus (Ebolavirus); MBGV, Marburg marburgvirus (Marburgvirus); LLOV, Lloviu virus (Cuevavirus); BoDV-1, Borna disease virus 1 (Bornavirus); NYMV, Nyamanini virus (Nyavirus). Their GenBank accession numbers are listed in Supplementary Table S1. The numbers on the left and right of the sequences indicate the amino acid positions in respective L proteins.
