Figure 2.
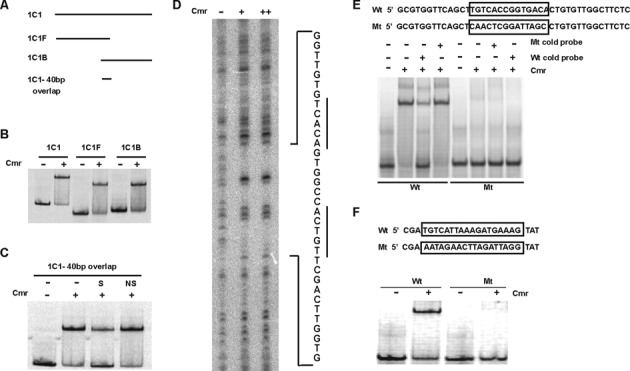
Mapping Cmr's DNA binding sequence in SELEX fragment 1C1. (A) Schematic of DNA fragments used to map Cmr binding site in 1C1. 1C1, full-length fragment (185 bp); 1C1F, forward fragment (101 bp); 1C1B, back fragment (124 bp). There is a 40 bp overlap between 1C1F and 1C1B, shown by 1C1–40 bp overlap. (B) EMSA, showing in vitro DNA binding ability of Cmr with 1C1, 1C1F and 1C1B as probes, in the absence or the presence of 300 nM His-Cmr recombinant protein, as indicated (C) EMSA showing DNA binding ability of His-Cmr with the 40 bp overlapping DNA fragment between 1C1F and 1C1B, as probe. Lane denoted ‘S’ was supplemented with 500× unlabeled 1C1 40 bp fragment as specific competitor DNA; Lane denoted ‘NS’ was supplemented with a 40 bp DNA fragment upstream of Rv1624c, as non-specific competitor DNA. (D) DNase I footprinting assay with His-Cmr using 1C1 DNA as template. 1C1 DNA was digested in the absence of Cmr (lane indicated ‘−’) or in the presence of 1.5 μM (lane ‘+’) or 3 μM Cmr (lane ‘++’). Sequence of the region protected from DNase I digestion, in the presence of Cmr, is shown adjacent to the figure. The palindromic half sites in the sequence are underlined. (E) Confirmation of Cmr's DNA binding motif recognition specificity, using mutational analyses. Sequence mutation of Cmr binding motif in 1C1 40 bp overlap region. The boxed WT (Wt) sequence was replaced as boxed in the mutant sequence (Mt). EMSA showing DNA binding ability of His-Cmr with WT and mutated motif sequences in 1C1 overlap region, in the absence (−) or presence (+) of 300 nM Cmr. 500× cold competition (Wt or Mt DNA) was used wherever indicated. (F) Sequence mutation of Cmr-binding motif in Rv1265 promoter. The underlined sequence in Rv1265 WT promoter (Wt) was replaced as shown in the mutant sequence (Mt). EMSA showing binding ability of His-Cmr with Rv1265 WT and mutated promoter sequences in the absence (−) or presence (+) of 900 nM Cmr.
