Figure 2.
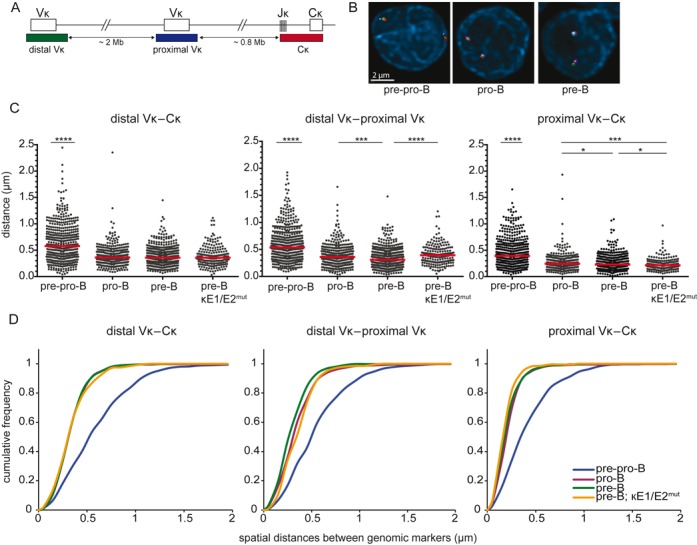
Igκ locus contraction in pro-B and pre-B cells is independent of iEκ. (A) Schematic representation of the murine Igκ locus including the bacterial artificial chromosome clones used as 3D FISH probes: distal Vκ – Alexa488, proximal Vκ – Cy5, Cκ – Alexa568. The indicated distances separating each of the 3 probes and their positions within the Igκ locus were determined from the Ensembl mouse genome database. (B) Representative images of the Igκ locus in the 3 B-cell subsets. (C) Scatter plots show the distances between distal Vκ, proximal Vκ, and Cκ regions in cultured E2A−/− pre-pro-B, Rag1−/− pro-B, Rag1−/− Igμ pre-B and Rag2−/− κE1/E2mut pre-B cells. For each condition 2–3 mice were used and at least 200 alleles were analyzed per population. Red horizontal lines represent median distances. The non-parametric Mann-Whitney test was used to calculate significance levels: *, P < .05; ***, P < .001; ****, P < .0001. (D) Cumulative frequency plots showing the distribution of spatial distances between the distal Vκ, proximal Vκ and Cκ region probes in the 4 progenitor-B cell subsets.
