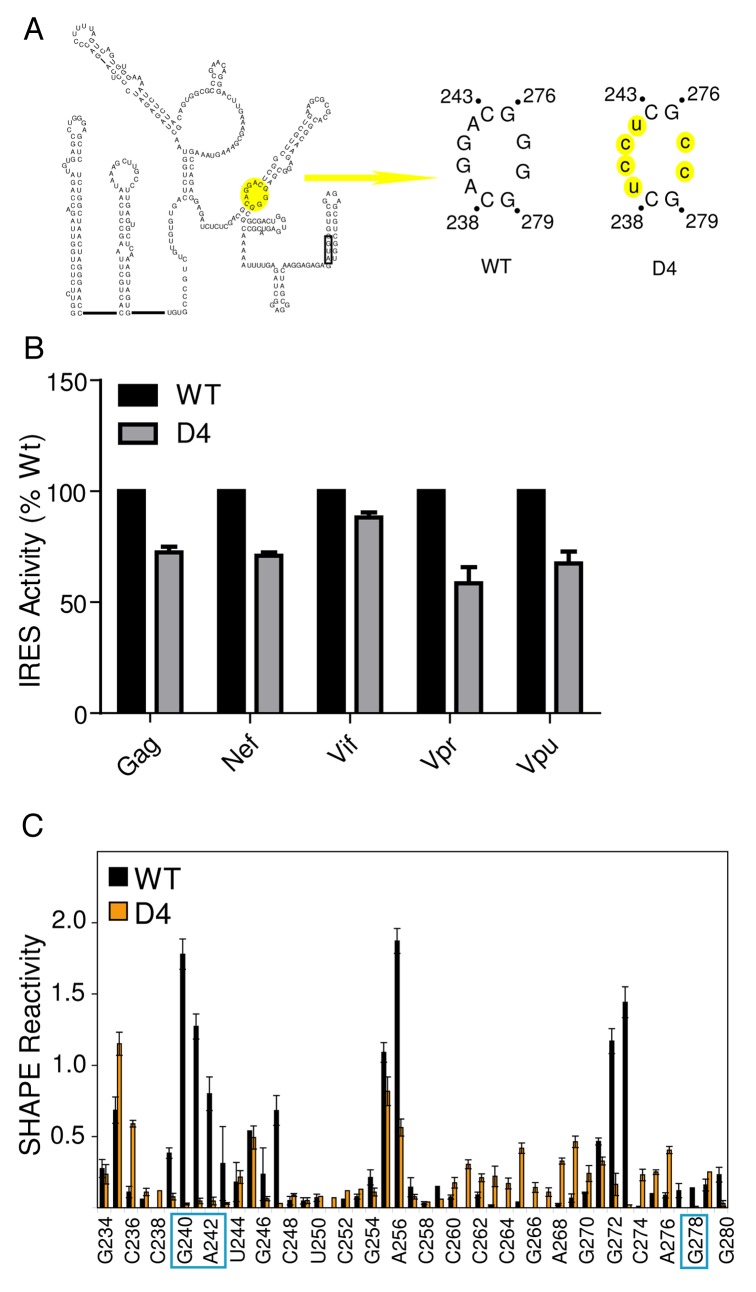
Figure 4. Regions within the DIS are important for IRES activity in HIV-1 transcripts. A. Schematic diagram of one of two proposed secondary structure models of the HIV-1 gag 5′ leader. The AUG hairpin model is presented.16,58 Base-pairing within the DIS domain I is proposed to be identical in each. The D4 mutation is highlighted in yellow in the AUG hairpin. WT denotes the common exon sequence naturally contained in all leader RNAs, and D4 denotes the mutation made in the leader RNAs. Mutated nucleotides are lower case and highlighted in yellow. B. IRES activity measured from Jurkat cells transfected with dual-luciferase DNA constructs containing WT and D4 sequences (as shown in panel A). IRES activity from each WT leader construct is set at 100%. C. Quantitated, normalized, and background-corrected modification data from at least two independent SHAPE probing experiments of the WT gag leader RNA and the D4 mutant RNA. The region surrounding the mutation is presented. The relative SHAPE reactivity is on the y-axis and the sequence is on the x-axis. Black bars denote reactivity of WT RNA, orange bars are reactivity of mutant RNA. The locations of the mutated nucleotides are boxed.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.