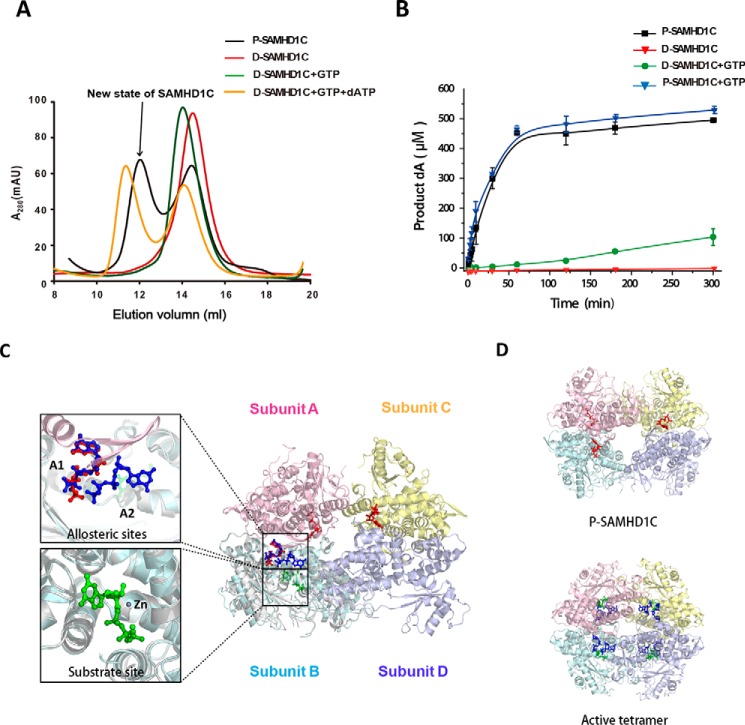
FIGURE 1.
The new state of SAMHD1C possesses a more efficient dNTPase activity than the dimer induced by GTP. A, size-exclusion column profiles of different states of SAMHD1C. P-SAMDH1C (purified SAMDH1 109–626) was purified from E. coli without nucleotides in the solution. D-SAMDH1C was obtained by dialysis of P-SAMDH1C into buffer (20 mm Tris-HCl, pH 8.0, with 200 mm NaCl, 5 mm MgCl2, and 0.5 mm TCEP) overnight to remove the nucleotides bound to the protein in vivo. mAU, milliabsorbance units. B, P-SAMDH1C has a more efficient dNTPase activity than the induced dimer of D-SAMDH1C+GTP. The amount of dA generated at each indicated time point, as described under “Experimental Procedures,” is shown. The experiment was repeated three times (n = 3). The error bars indicate the S.D. of three replicates. C, graphic representation of the new form of SAMHD1C. One subunit from 4QFX overlapping the subunit B is shown to indicate the positions of the two allosteric sites (A1, A2) and a substrate site. Subunits A, B, C, and D are colored pink, light blue, light yellow, and light purple, respectively. Three GTP activators occupying the A1 sites are shown as red sticks. In 4QFX, two dGTP molecules occupying the allosteric sites (A1, A2) are colored deep blue, and a dGTP molecule occupying the substrate site is in green. D, overall structure comparison of the active tetramer 4QFX and our structure.