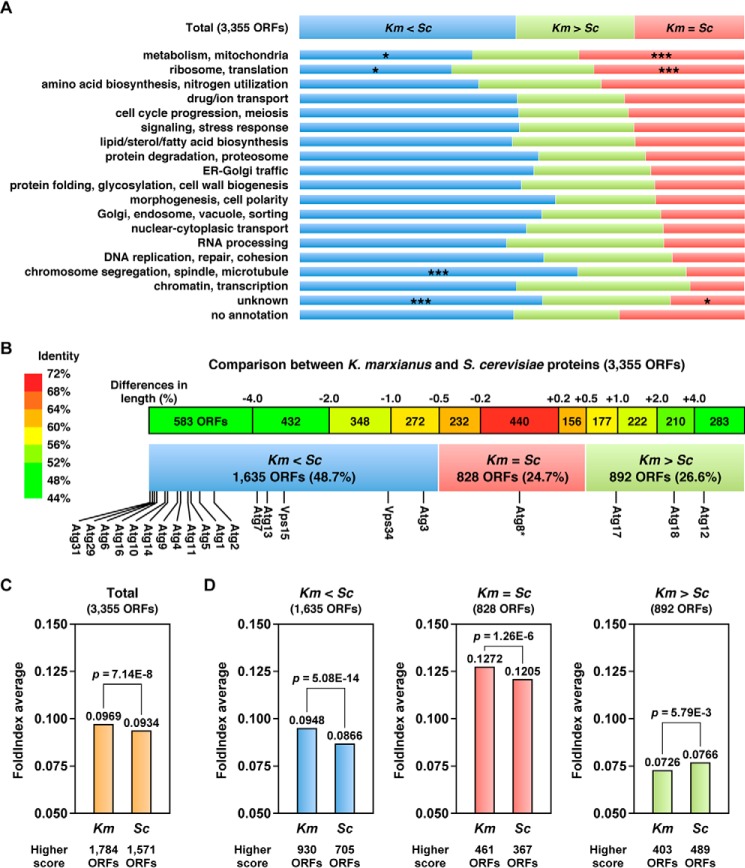
FIGURE 1.
Bioinformatic analyses of the comprehensive genome sequences of K. marxianus and S. cerevisiae. A, comparable ORFs aligned using the BLAST server were classified into three subgroups according to their relative lengths (see also supplemental Table 2). The comparable ORFs (total 3,355 ORF pairs) were annotated according to their functions. Asterisks, significantly fewer than in total ORFs (p < 0.005); triple asterisks, significantly more than in total ORFs (p < 0.005). B, almost half of all K. marxianus proteins are shorter than their S. cerevisiae counterparts. Km < Sc, the K. marxianus proteins are >0.5% shorter than the S. cerevisiae counterparts (1,635 ORFs in blue); Km = Sc, the K. marxianus proteins are nearly the same length as the S. cerevisiae counterparts (828 ORFs in red); Km > Sc, the K. marxianus proteins are >0.5% longer than the S. cerevisiae counterparts (892 ORFs in green). Identities of the comparable ORFs (3,355 ORF pairs) from A were calculated using the LALIGN server. Asterisk, mature form of Atg8. C, unfoldability scores of all comparable K. marxianus and S. cerevisiae ORFs (3,355 ORF pairs) were assessed using the FoldIndex server (see also supplemental Table 3). Mean FoldIndex scores were analyzed statistically. D, mean FoldIndex scores of three subgroups in B were analyzed as in C.