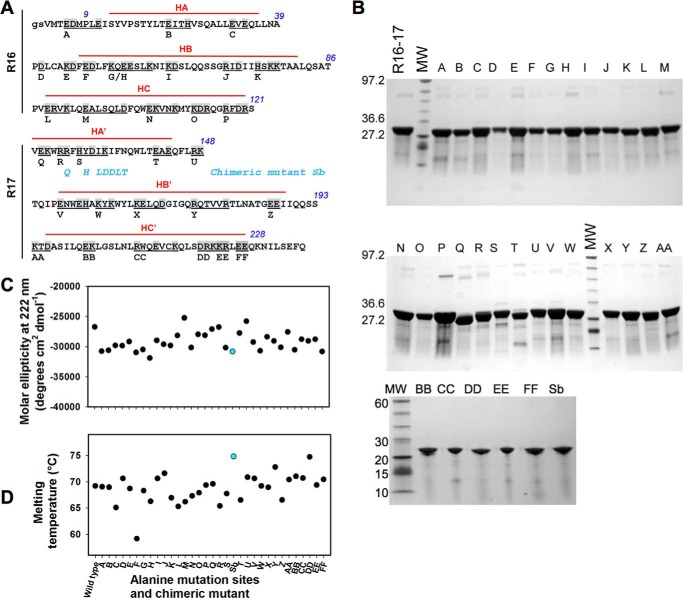
FIGURE 1.
Biochemical characterization of the dystrophin R16–17 fragment and mutants. A, for alanine-scanning mutagenesis, all charged residues of dystrophin R16–17 were changed to alanine in groups of 1 to 5 amino acids (common value of 2–3 amino acids). This grouping encompassed 74 residues (shown in gray), resulting in 32 variants, A through FF. The capital letters are the R16 and R17 sequence according to Koenig and Kunkel (38); the gs italicized letters are residues present due to cloning constraints. The three helices of each repeat are indicated in red lines and noted as HA, HB, and HC for repeat 16 and HA′, HB′, and HC′ for repeat 17. The residues not marked by red lines are those involved in the loops between successive helices. There is no interruption of the HC and HA′ of repeats 16 and 17, respectively, with these two helices forming a common helix with the residues in the junction involved in the so-called linker. The chimeric mutant Sb not corresponding to alanine-scanning mutagenesis is shown: the blue residues are those substituted from the utrophine sequence. B, the native fragment of dystrophin R16–17, the 32 alanine mutants, and the chimeric Sb mutant were produced and purified. The fragments appeared at the expected molecular masses of ∼22,700 Da and at a high degree of purity. MW, molecular weight standards. C, molar ellipticity values at 222 nm as measured from CD spectra for the wild-type dystrophin R16–17 and all the mutants. The chimeric mutant value appears in blue. D, melting temperatures of wild-type dystrophin R16–17 and all the mutants as obtained by heating from 15 to 85 °C and followed by CD at 222 nm. The chimeric mutant value appears in blue.