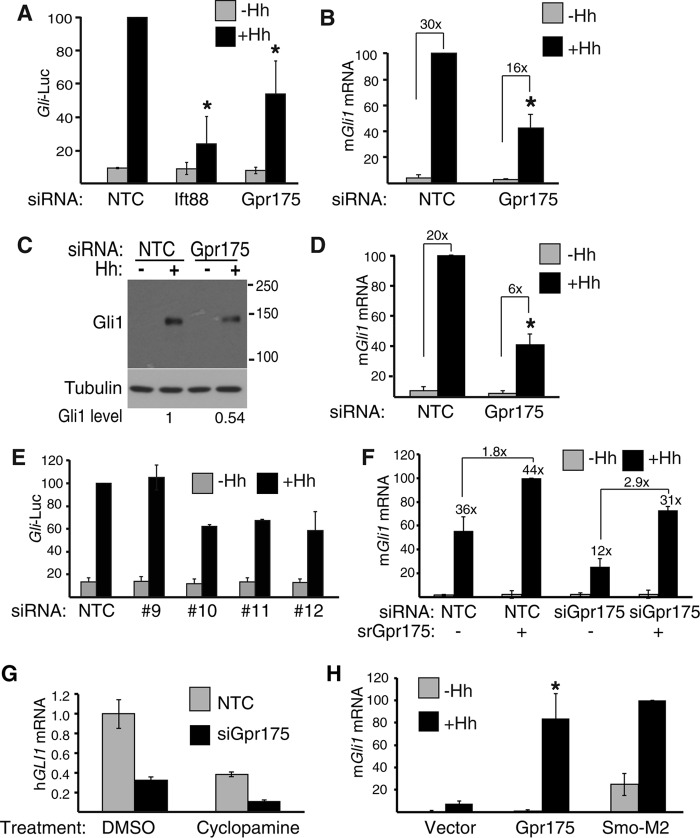
FIGURE 2.
Gpr175 depletion impairs Hh signaling. A, percentage Gli-luciferase (Gli-Luc) activity remaining after transfection with siNTC (non-targeting control), siIft88 (positive control), and siGpr175 (all pools of 4 siRNAs) in S12 cells with (black) or without Hh (gray). Data were normalized to 100% for siNTC + Hh and the mean ± S.D. of four individual triplicate experiments are shown (*, p ≤ 0.05). B, endogenous murine Gli1 expression in S12 cells by qRT-PCR following Gpr175 depletion with (black) or without Hh (gray) and normalized to mRpl19 and siNTC -Hh control (n = 3; *, p ≤ 0.05). The mean fold-induction by Hh for each siRNA is indicated above each pair of bars. C, representative Western blot of cells treated as in B and probed with a Gli1-specific monoclonal antibody. Quantitation of three such blots normalized to tubulin revealed a 54.3 ± 20.5% decrease in Gli1 protein (p = 0.015). D, as in B but using mouse embryonic fibroblasts (n = 3; *, p ≤ 0.05). E, three of four individual Gpr175 siRNAs of the pool suppress signaling; n = 3 experiments performed as in A. F, rescue of murine Gpr175 knockdown (by pool of siRNAs #10–12) with siRNA-resistant mouse Gpr175-LAP5 (srGpr175) in C3H10T1/2 cells; mGli1 induction by Hh was monitored by qRT-PCR (mean ± S.D. of 3 independent experiments). Numbers above the black bars indicate the fold-increase over −Hh for that treatment; fold-increase induced by Gpr175 over empty vector (both +Hh) is indicated above the connecting lines. G, human medulloblastoma Daoy cells were depleted of GPR175 and endogenous hGLI1 levels were monitored in the absence of Hh stimulation (because this cell line is constitutively active) with and without 16 h treatment with 2 μm Smo inhibitor cyclopamine. hGLI1 mRNA levels were normalized to hRPL19. H, overexpression of Gpr175 enhances Hh signaling. Endogenous mGli1 expression by qRT-PCR following transfection of empty vector only, Gpr175-LAP5 or SMO-M2 with (black) or without Hh (gray) in C3H10T1/2 cells. Data were normalized to the housekeeping gene mRpl19 mRNA and expressed as percentage of SMO-M2 + Hh. Mean ± S.D. of three individual experiments are shown (*, p ≤ 0.05). Error bars in all panels represent mean ± S.D. of at least three independent triplicate experiments.