Figure 4. Heavy and light chain of the VRC01 lineage exhibit multi-clade phylogenetic organization and concordance.
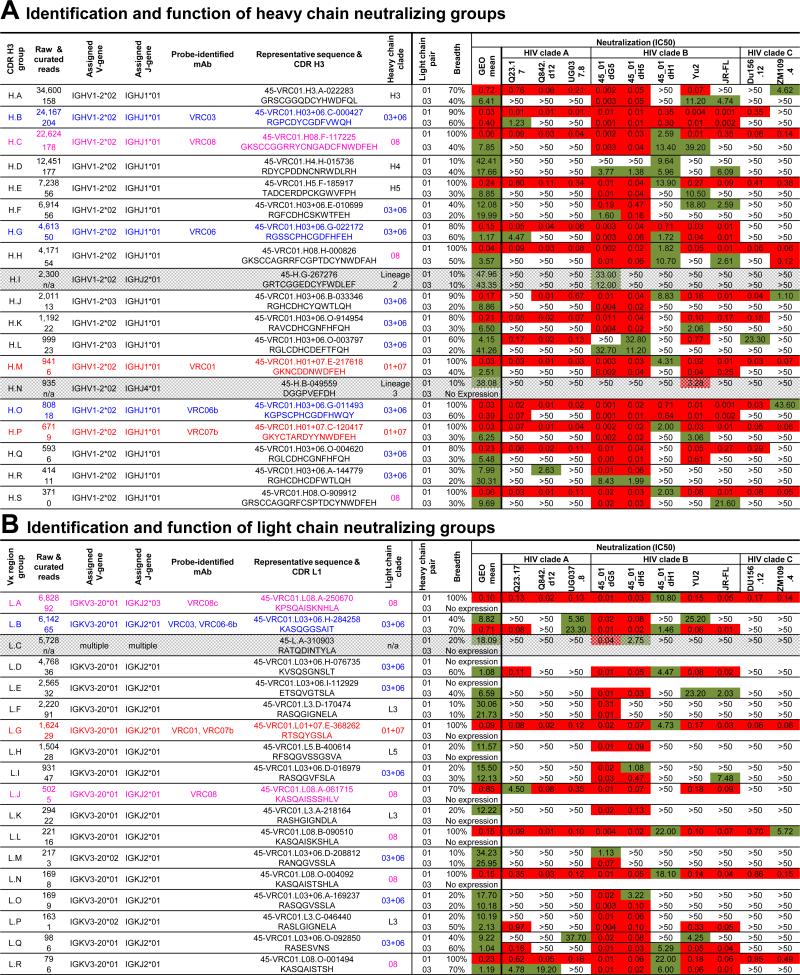
(A) Heat map showing pairwise sequence identities for heavy chains of probe-identified antibodies and validated neutralizing heavy chain sequences sorted by overall prevalence (among final curated sequences). Groups with high identity and similar prevalences were merged into clades, shown at left.
(B) Heat map for light chains of probe-identified antibodies and validated neutralizing light chain sequences, produced as in (A).
(C) Maximum-likelihood phylogenetic trees for heavy chain (left) and light chain (right) sequences. The clades described in (A) and (B) can be clearly seen in the structure of the trees, which have similar overall topology. Temporal prevalence (middle) is charted for each clade as the fraction of unique sequences with an in-frame junction and no stop codons (but without manual curation) at each time point which are assigned to that clade. For clades 01+07, 03+06, and 08, which are anchored by probe-identified antibodies, the correlation of heavy and light chain-temporal prevalence is shown (boxes). The average divergence for each clade at each time point is calculated from curated sequences.
See also Figure S4.