Abstract
Modular polyketide synthases (type I PKSs) in bacteria are responsible for synthesizing a significant percentage of bioactive natural products. This group of synthases has a characteristic modular organization, and each module within a PKS carries out one cycle of polyketide chain elongation; thus each module is “non-iterative” in function. It was possible to predict the basic structure of a polyketide product from the module organization of the PKSs, since there generally existed a co-linearity between the number of modules and the number of chain elongations. However, more and more bacterial modular PKSs fail to conform to the “canonical rules”, and a particularly noteworthy group of non-canonical PKSs is the bacterial iterative type I PKSs. This review covers recent examples of iteratively-used modular PKSs in bacteria. These non-canonical PKSs give rise to a large array of natural products with impressive structural diversity. The molecular mechanism behind the iterations is often unclear, presenting a new challenge to the rational engineering of these PKSs with the goal of generating new natural products. Structural elucidation of these synthase complexes and better understanding of potential PKS-PKS interactions as well as PKS-substrate recognition may provide new prospects and inspirations for the discovery and engineering of new bioactive polyketides.
Keywords: Biosynthesis, Natural products, Polyketides, Type I PKS, Iterative PKS
Introduction
Polyketides are a highly diverse group of natural products, which are biosynthesized from short acyl-CoA units by polyketide synthases (PKSs) (Hopwood 1997; Staunton and Weissman 2001; Hertweck 2009). The polyketide natural products represent an important source of novel therapeutics, and research of polyketides has led to the discovery of an enormous variety of molecular structures with a wide range of biological activities. Many of these have found use as new pharmaceuticals, such as antibiotics, immunesuppressants, antiparasitics, hypolipidemics, and anti-tumoral agents (Fischbach and Walsh 2006).
Over the past two decades, the biosynthetic gene clusters for a broad range of polyketide natural products have been identified and characterized, and the studies of structures, activities and mechanisms of PKSs have led to strategies in rational engineering and combinatorial biosynthesis, which have further expanded the polyketides’ structural diversity (Khosla 2000; Wilkinson et al. 2003). Bacterial modular PKSs (type I) are particularly attractive targets for this purpose, due to their defined modular organization, remarkable versatility, and amenability for pathway engineering (Shen and Thorson 2012; Watanabe 2008; Weissman and Leadlay 2005; Williams 2013; Winter and Tang 2012; Zabala et al. 2012). In general, a PKS module consists of individual domains in charge of selecting, fusing, and processing the building blocks. The substrates for chain initiation and extension are selected and activated by an acyl transferase (AT) domain, and transferred to the 4’-phosphopantetheinylated acyl carrier protein (ACP) domain. The ketosynthase (KS) domain catalyzes the decarboxylative Claisen-like condensation between the substrate and the growing polyketide, forming a carbon-carbon bond between the alpha carbon of the extender unit and the thioester carbonyl of the ACP-bound acyl chain. While a PKS module requires a minimum of these three domains to function, other domains are commonly present which serve to further tailor the nascent polyketide: the ketoreductase (KR), dehydrogenase (DH), and enolreductase (ER) domains function sequentially to reduce the β-keto group into a fully saturated acyl chain (Fischbach and Walsh 2006). Because PKS modules vary in the presence of these β-keto-processing domains and thus in the degree to which the β-keto is reduced, PKSs generate a vast diversity and complexity of polyketide products (Chan et al. 2009) (Fig. 1).
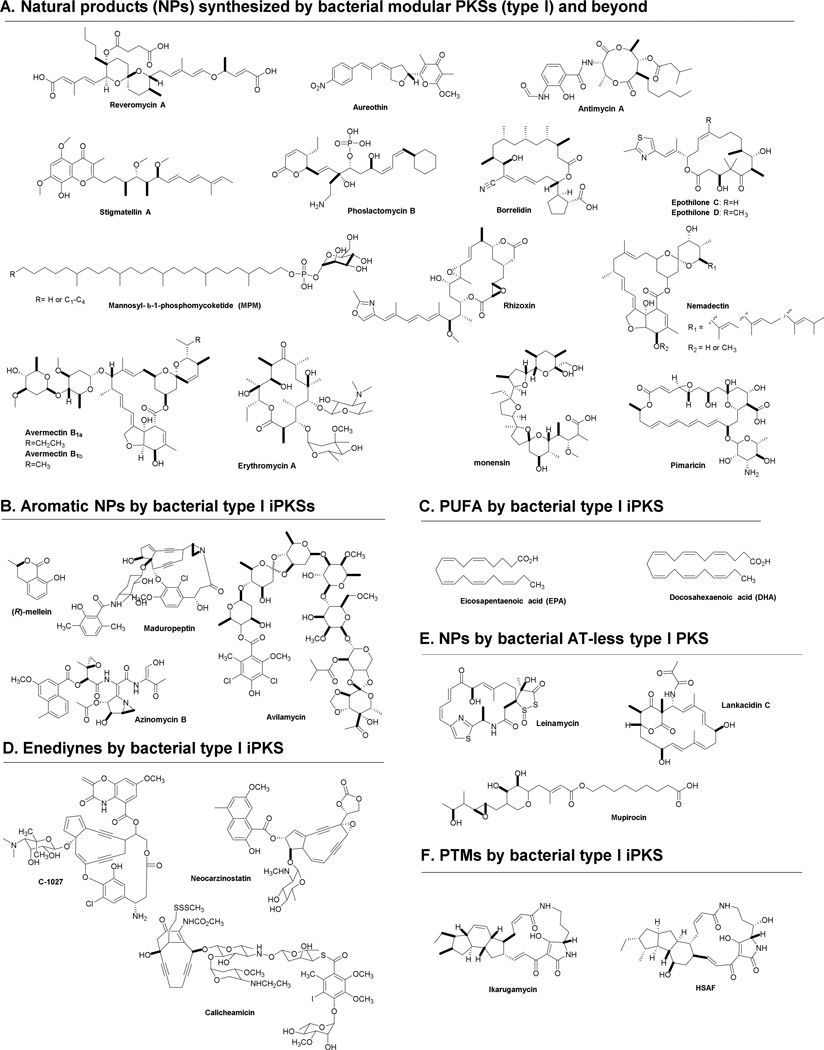
Fig. 1.
Examples of natural products synthesized by bacterial modular PKSs (type I) that are discussed in this review.
PKSs are classified according to the structural organization of their functional domains. Type I PKSs consist of a single large polypeptide chain housing multiple different domains capable of catalyzing different reactions. Such “multimodular” PKSs are found in both bacteria and fungi. Type II PKSs are usually found in bacterial systems and are made up from separate dissociable proteins, each serving a different function during polyketide synthesis (Zhang and Tang 2009). Typical products of type II PKSs are aromatic metabolites, although exceptions are known (Gaitatzis et al. 2002). Type III PKSs utilize acyl-coenzyme A (CoA) thioesters directly, rather than depending on extender units tethered to an ACP. Type III PKSs also produce aromatic metabolites and are mainly distributed in the plant kingdom, but occasionally occur in bacteria (Moore and Hopke 2001). While these three classifications of PKSs are useful generalizations, nature does not limit itself to only three rigidly-defined methods for generating polyketide metabolites (Muller 2004). In this review, we will focus our discussion on examples in which bacterial modular PKSs (type I) synthesize polyketides in a non-canonical iterative manner, which combines the two traits once thought to be mutually exclusive to fungal or to bacterial systems (Fig. 1).
Type I PKS
Type I PKSs are often further subdivided into two major groups: the iterative (fungal) type I PKSs and non-iterative (bacterial) type I PKSs (Cox 2007; Hertweck 2009; Hopwood 1997; Smith and Tsai 2007; Staunton and Weissman 2001). A fungal iterative PKS (iPKS) is usually a single-module protein consisting of a single set of functional domains. Although containing only a single module, a fungal iPKS is nevertheless capable of conducting multiple rounds of chain extension and β-keto processing. The varied reduction level of the β-keto during each round of elongations is the most intriguing feature of this group of PKSs (Crawford and Townsend 2010; Fujii 2010; Kennedy et al. 1999). These iPKSs are largely confined to fungal systems, and thus will not be discussed in this review. Bacterial type I PKSs, in contrast, include multiple sequential modules, each of which contain the needed extending and tailoring domains, and operate in a modular fashion with each module being responsible for only one round of chain elongation and subsequent β-keto processing before passing the nascent polyketide onto the downstream module which carries out another round of chain extension and processing. Examples of such modular type I PKSs include those responsible for the biosynthesis of the macrolides such as erythromycin (Cortes et al. 1990; Donadio et al. 1991; Smith and Tsai 2007) and avermectin (Omura et al. 2001). These bacterial type I PKSs thus carry out each successive chain extension cycle by a different set of active sites (domains) housed in separate modules, and one or more such modules may exist on each multifunctional polypeptide. Importantly, in most modular type I PKSs, each active site is used only once; thus the modular type I PKSs are said to be non-iterative. This linear mechanism was found in the biosynthesis of numerous bacterial polyketides by type I PKSs (Broadhurst et al. 2003; Challis 2008; Khosla et al. 2009; Moss et al. 2004; Sherman 2005; Winter et al. 2011).
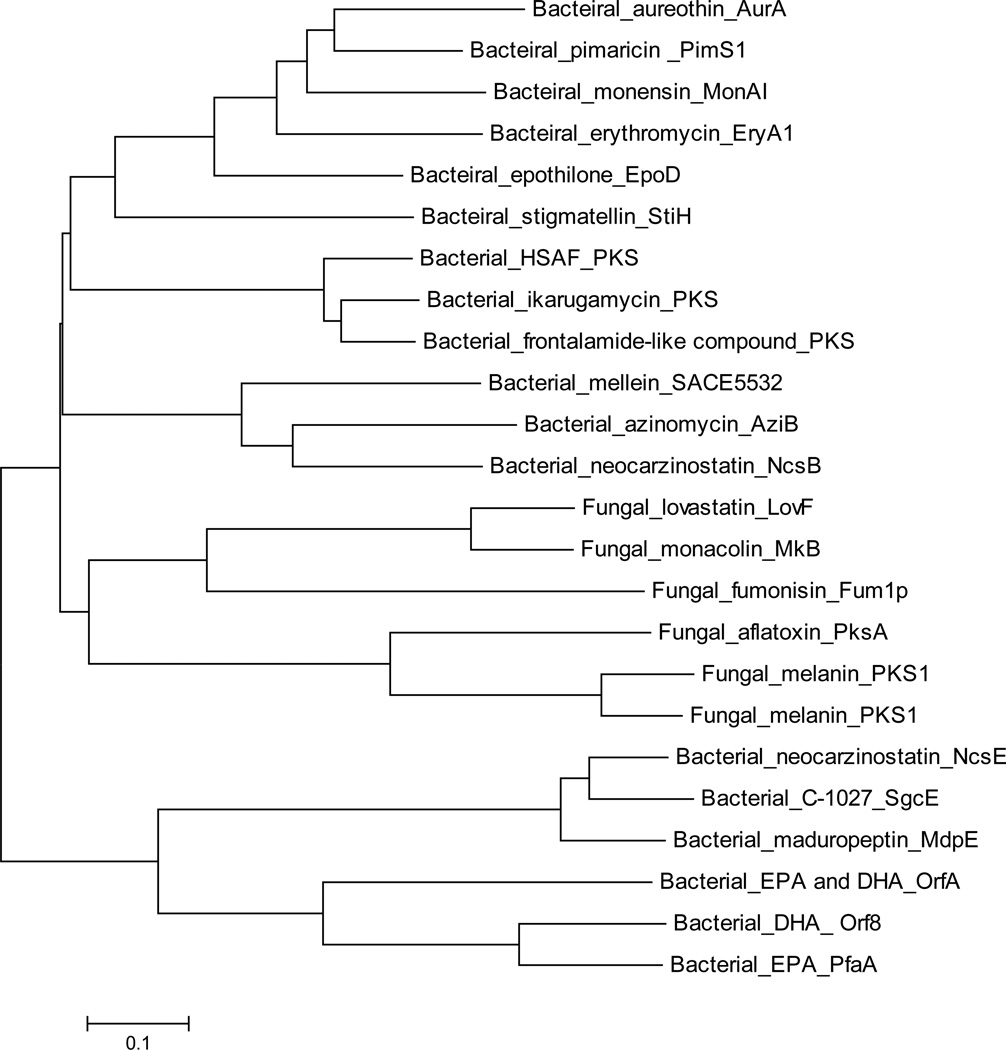
Despite the “canonical” organization and co-linearity found in the majority of bacterial type I PKSs, non-canonical examples have continued to emerge from bacteria (Fig. 1) (Fisch 2013; Shen et al. 2007). These non-canonical type I PKSs possess the basic modular organization, but often include unusual arrangements of the functional domains. They exhibit a phylogenetic distance from either fungal iterative type I PKSs or bacterial non-iterative type I PKSs, making them fall into five new subgroups, including bacterial type I iPKS for aromatic compounds (Zhang et al. 2013), polyunsaturated fatty acid (PUFA) synthases (Kaulmann and Hertweck 2002; Metz et al. 2001), enediyne synthases (Liu et al. 2002; Van Lanen et al. 2008), polycyclic tetramate macrolactams (PTM) synthases (Blodgett et al. 2010; Li et al. 2014) and some trans-AT type I PKSs (Cheng et al. 2003; El-Sayed et al. 2003; Gay et al. 2014; Musiol and Weber 2012; Piel 2010). These phylogenetic studies have shown the proposed evolutionary relationship underlying the catalytic diversity of PKSs (Jenke-Kodama et al. 2005; Zhang et al. 2013). The bacterial aromatic iPKSs and PTM-type PKSs make up two independent clades that lie close to each other and sister to canonical type I PKSs, but are phylogenetically distant from the clades containing two other types of bacterial iPKSs, the enediyne synthases and PUFA synthases, as well as the clades containing fungal non-reducing PKSs (NR-PKSs) and highly reducing PKSs (HR-PKSs) (Crawford and Townsend 2010; Liu et al. 2015b) (Fig. 2). The phylogenetic differences and relationships between the subgroups in type I PKSs has given rise to the development of a rapid PCR approach to specifically access the genes encoding these enzymes (Jia et al. 2006; Zhao et al. 2008).
Fig. 2.
Phylogenetic analysis of different types of PKSs based on the sequence of KS domains. Enzymes and their GenBank accession numbers are listed as follows: bacterial aromatic iPKS (3 sequences, including AziB for azinomycin biosynthesis, ABY83164.1; NcsB for neocarzinostatin biosynthesis, AAM77986.1; SACE5532 for mellein biosynthesis, YP_001107644.1); PTM type iPKSs (3 sequences, including PKS for HSAF biosynthesis, EF028635.2; PKS for ikarugamycin biosynthesis, KF954512.1; PKS for frontalamide-like compound biosynthesis from Streptomyces albus J1074, ABYC01000481); modular PKSs (6 sequences, including PimS1 module 1 for pimaricin biosynthesis, CAC20931.1; MonAI: module 1 for monensin biosynthesis, AAO65796.1; EryA1: module 2 for erythromycin biosynthesis, YP_001102988.1; AurA for aureothin biosynthesis, CAE02602.1; EpoD for epothilone AAF26921.1; StiH for stigmatellin CAD19092.1); HR-PKSs (3 sequences, including Fum1p for fumonisin biosynthesis, AAD43562.2; LovF for lovastatin biosynthesis, AAD34559.1; MkB for monacolin biosynthesis, ABA02240.1); NR-PKS (3 sequences, including PksA for aflatoxin biosynthesis from Aspergillus flavus, AAS90093.1; PKS1 for melanin biosynthesis from Colletotrichum lagenaria, BAA18956.1; PKS1 for melanin biosynthesis from Glarea lozoyensis, AAN59953.1); enediyne synthases (3 sequences, including SgcE for C-1027 biosynthesis, ZP_11383500.1; MdpE for maduropeptin biosynthesis, AAQ17110.2; NcsE for neocarzinostatin biosynthesis, AAM78012.1); PUFA synthases (3 sequences, including PfaA for EPA biosynthesis from Photobacterium profundum, AAL01060.1; Orf8 for DHA biosynthesis from Moritella marina, BAA89382.2; OrfA for EPA and DHA biosynthesis from Schizochytrium AAK72879.2). Similar sequences were aligned with ClustalW and the tree shown was generated using the MEGA 5.0).
Non-canonical bacterial type I PKS
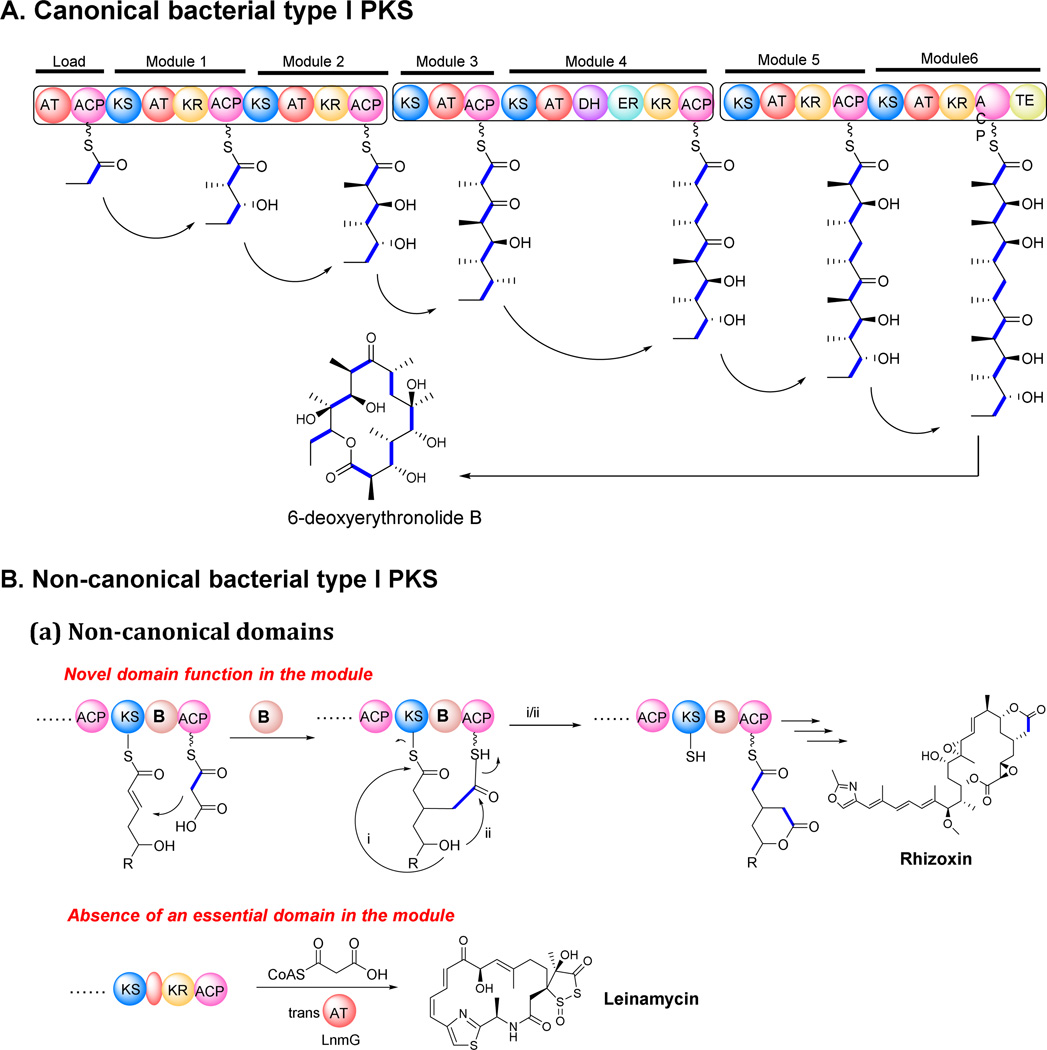
Bacterial type I PKSs were originally thought to be rigidly modular and co-linear with the core structure of final products (Fig. 3A), so that the basic structure of an unknown compound produced by this type of PKSs could be deduced from bioinformatics analysis of the PKS genes. Due to the high level of amenability, type I PKS systems have the potential to be engineered to make novel analogues of known metabolites. Indeed, new compounds have been accessed by re-programming type I PKS systems in a number of ways, such as loading module swapping, module deletion or insertion, and domain swapping or inactivation (Xu et al. 2013).
Fig. 3.
Examples of canonical bacterial type I PKSs and non-canonical bacterial type I PKS. KS, ketosynthase; AT, acyltransferase; DH, dehydratase; KR, ketoreductase; ACP, acyl carrier protein; TE, thiolesterase; C-MeT, C-methyltransferase; ER, enoyl reductase; B, branching domain.
However, many deviations from the “canonical rule” have been discovered in bacterial type I PKSs in recent years. Broadly speaking, there are two types of deviations: non-canonical domains and non-canonical modules. Non-canonical domains include presence of unusual domains, absence of domains for required functions, loss of domain specificity, and novel domain functions and domain organization (Katz 2009). For example, the newly discovered branching domain acetylates an enzyme-bound enoyl moiety by means of a Michael-like conjugate addition, resulting in a β-branch instead of an elongated backbone in rhizoxin biosynthesis (Fig. 3B) (Bretschneider et al. 2013). Trans-acting domains are discrete enzymes that act only at specific points during the synthesis of nascent acyl chains, remaining physically separate from the usual modifying domains in the modular PKSs. Examples include the trans AT in AT-less PKS systems such as leinamycin synthase (Fig. 3B) and kirromycin synthase (Cheng et al. 2002; Cheng et al. 2003; Musiol et al. 2011), the discrete KR domain SiaM from the SIM7248 biosynthetic pathway and AntM from the antimycins biosynthetic pathway (Zou et al. 2013; Liu et al. 2015a), and the discrete DH domain in the biosynthesis of phoslactomycin (Chen et al. 2012; Palaniappan et al. 2008). The reveromycin PKS is another example where the genetic organization of the biosynthetic gene cluster and the corresponding proteins are not co-linear (Takahashi et al. 2011). The analysis of the reveromycin PKS shows that it is possible that after translation, RevC protein (containing module 1–3), associates with RevA (module 4–6) instead of RevD, whose gene is located next to and downstream of RevC, to bring the set of contiguous domains together to form functional modules. Therefore, even when the PKS genes are not present on the chromosome in a co-linear arrangement, the gene products form a specific head-to-tail complex through which the polyketide chain is processed in a programmed fashion and biosynthesis occurs in a co-linear fashion.
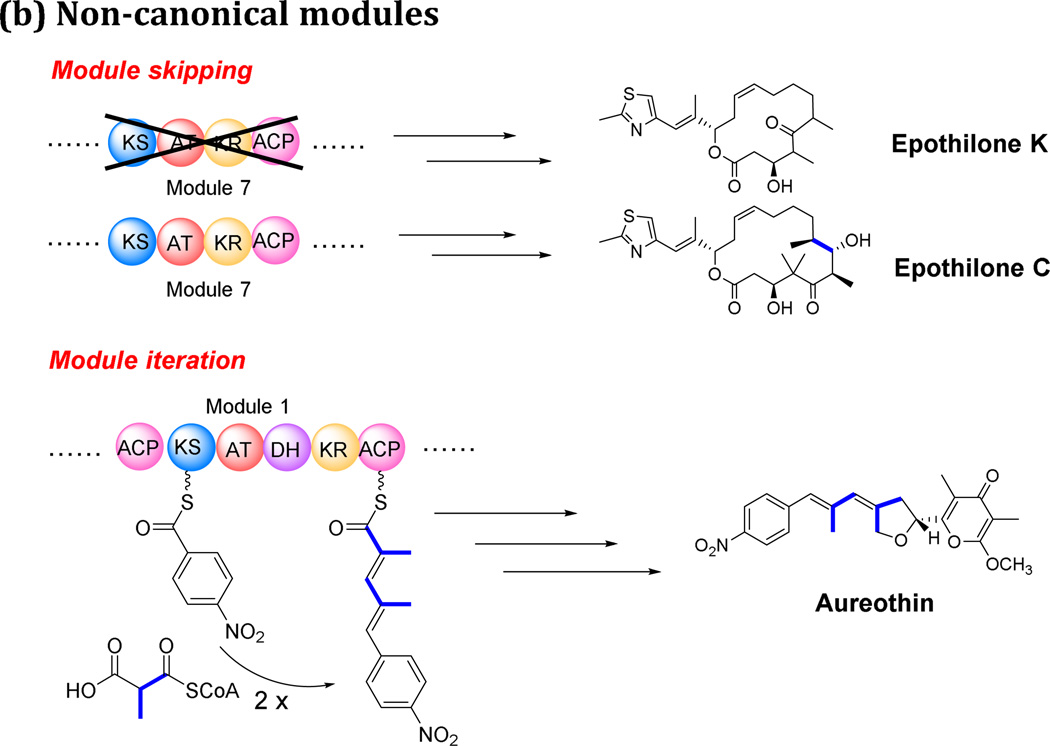
Non-canonical domains affect the co-linearity mainly in the modification of the backbone, such as generating unpredictable reduction levels of the β-keto group. But the non-canonical modules usually result in changing the size of the backbone in the final products. The most frequently encountered examples of such aberrations are the skipping of various single domains, or sometimes of entire modules (El-Sayed et al. 2003); conversely, individual modules may be used more than once, giving rise to the so-called “module stuttering”, which is one example of iterative behavior in bacterial PKSs (Moss et al. 2004). Such events lead to inconsistency between the genetic organization of the biosynthetic gene cluster and its corresponding proteins, and its ultimate PKS metabolite (Hertweck 2015).
A PKS which skips over modules usually does so in “programmed skips”, passing over extraneous functional domains or modules in a PKS for which there is no obvious need in the biosynthesis of the corresponding polyketide, such as occurs in the biosynthesis of mupirocin (El-Sayed et al. 2003). Truncated derivatives are also produced in trace amounts during the biosynthesis of avermectin and epothilone (Fig. 3B) (Julien et al. 2000; Molnar et al. 2000), suggesting the aberrant bypass of one or another module during biosynthesis which leads to unusual compounds.
Iterative use of one or more modules will often result in longer polyketide chains than those predicted by the co-linear model (Fig. 3B). This module iteration composes a large group of non-canonical bacterial type I PKS, and the iteration events can happen accidently or in a programmed manner. Besides those with a special domain organization, which already fall into new subclasses of type I PKSs (bacterial aromatic iPKS, PUFA synthases, enediyne synthases), there are still many extraordinary examples which do not conform to any existing classifications. In the following sections, we focus on the iteration events known to occur in bacterial type I PKSs and discuss potential mechanisms behind this phenomenon.
Iteratively used bacterial type I PKS
Bacterial iterative type I PKSs can be considered in two categories: entirely iterative PKSs (fungal iPKS-like) and partially iterative PKSs (module-iteration).
Entirely iterative bacteria PKSs
Bacterial type I iPKSs producing aromatic compounds
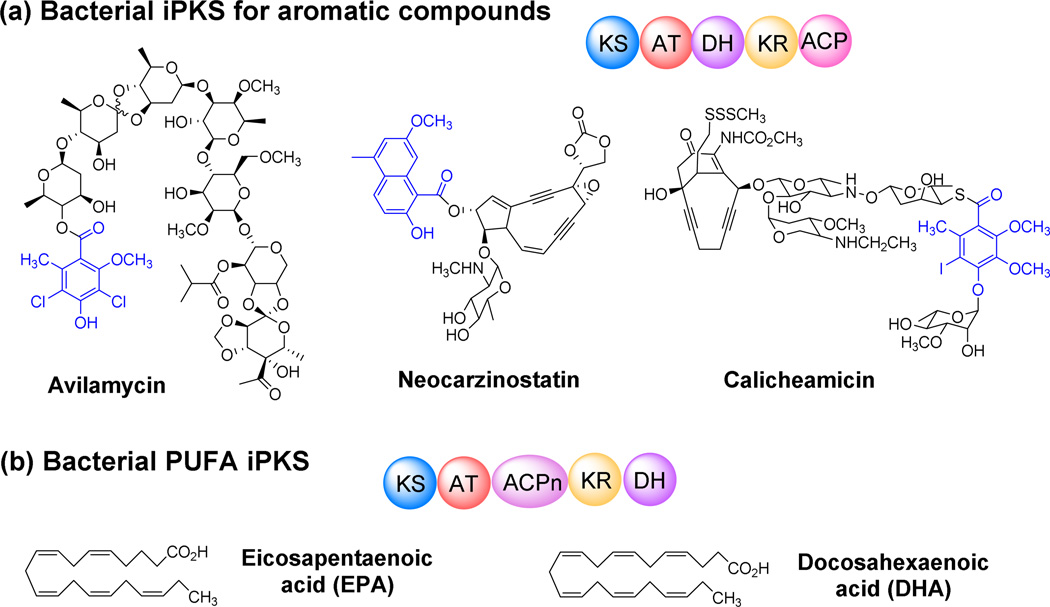
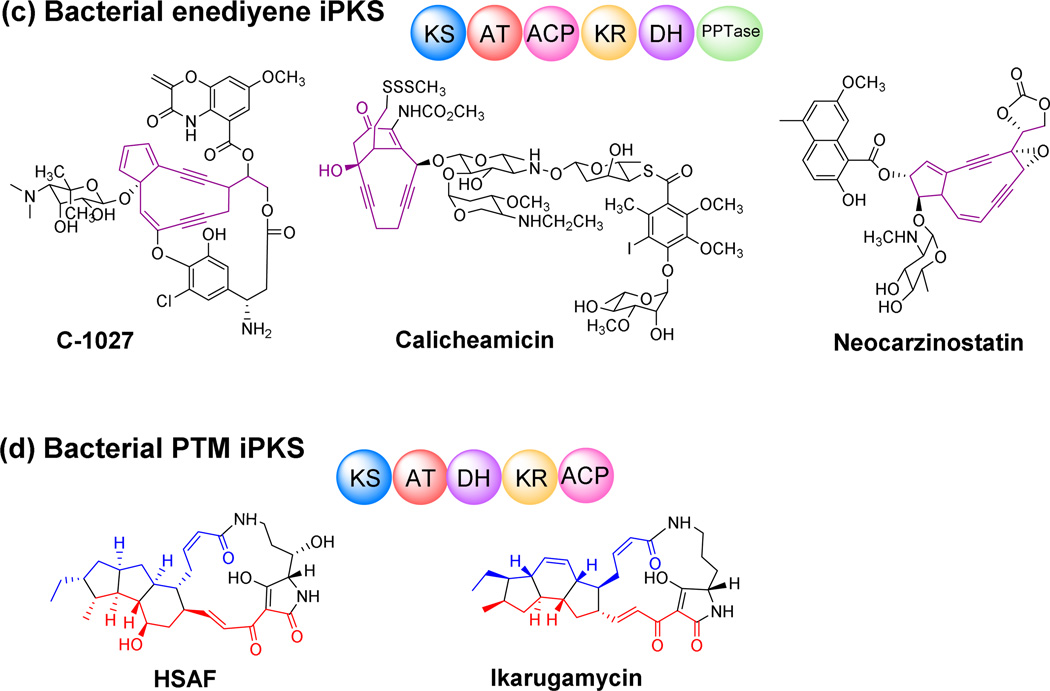
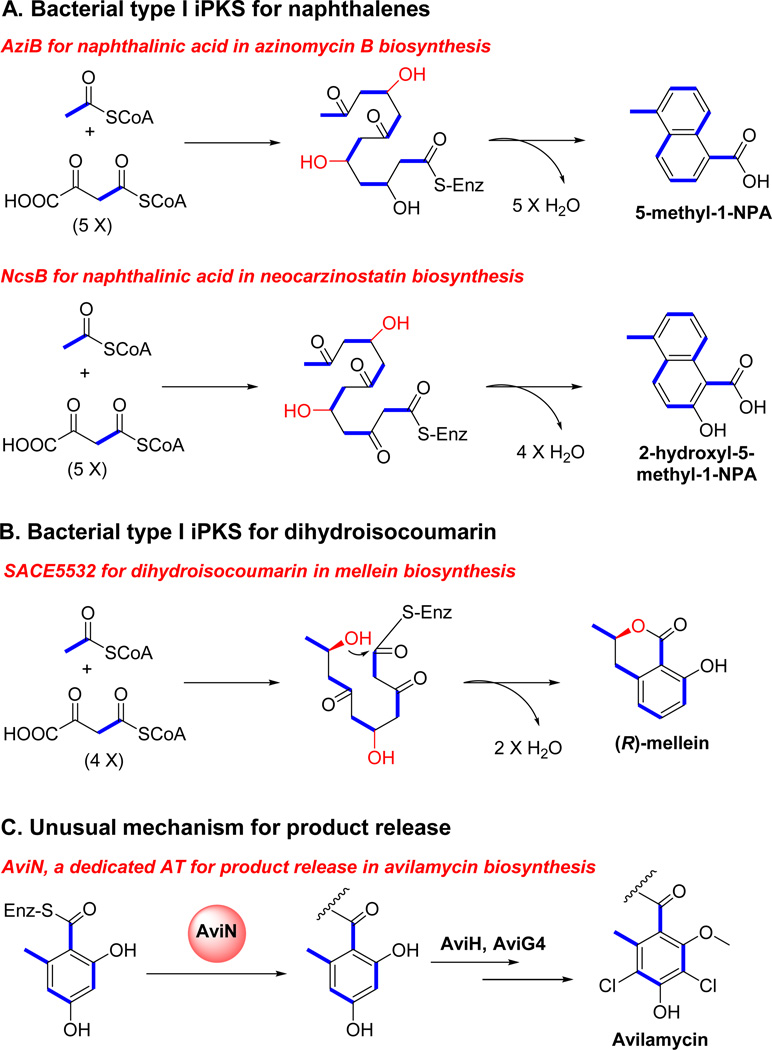
Most aromatic polyketides produced in bacteria are synthesized by type II (non-modular) PKSs. However in 1997, AviM, a single-module PKS, was found responsible for the biosynthesis of orsellinic acid, an phenolic moiety in avilamycin (Fig. 4a) (Gaisser et al. 1997). Heterologous expression provided experimental evidence for single-module AviM’s iterative synthesis of the aromatic moiety. Another example of bacterial iterative type I PKS for aromatic compounds is CalO5, which is involved in calicheamicin biosynthesis (Fig. 4a) (Ahlert et al. 2002). Besides the monocyclic aromatic polyketides, several bacterial type I PKSs were found to generate higher-order aromatic polyketides through iterative use of the same module, such as NcsB, which catalyzes the formation of a naphthalinic acid moiety in neocarzinostatin biosynthesis (Fig. 5a) (Zazopoulos et al. 2003). So far, bacterial type I iPKSs are associated with the biosynthesis of chlorothricin (Jia et al. 2006), maduropeptin (Van Lanen et al. 2007), polyketomycin (Daum et al. 2009), pactamycin (Ito et al. 2009), tiacumicin B (Xiao et al. 2011), azinomycin B (Ding et al. 2010) and (R)-mellein (Sun et al. 2012).
Fig. 4.
Domain organization of bacterial type I iPKSs and representative compounds produced by these synthases. The domain organizations of iterative type I PKSs are classified into type I iPKS for aromatic compounds, PUFA iPKS, enediyne iPKS, and PTM iPKS. ACPn, multiple ACPs; PPTase, 4’-phosphopantetheinyl transferase.
Fig. 5.
Iterative polyketide biosynthesis of aromatic compounds by bacterial type I iPKSs and an unusual product off-loading mechanism in avilamycin biosynthesis. The selective keto-reduction patterns are highlighted in red.
Bacterial aromatic iPKSs are typically organized in the order of KS-AT-DH-(KR)-ACP (Fig. 4a), and share high homology with each other, which is just like a common module of canonical type I PKSs. Interestingly, the module order of these bacterial iPKSs is also homologous to fungal iPKS, such as 6-MSA synthases (Fujii 2010), except the absence of C-methylation (CMeT) and enoylreductase (ER) domains.
Bacterial aromatic iPKSs contribute to the diversity of polyketides generally through their unusual domain functions, different mechanisms for polyketide off-loading and post-synthesis modifications (Fig. 5). For instance, the KR domain of most of the aromatic iPKSs contains a motif similar to the Leu-Asp-Asp motif of B-type ketoreductases, suggesting that all the KR domains of bacterial aromatic iPKSs specifically generate a D-hydroxy group by reduction of the β-keto group (Fig. 5) (Keatinge-Clay 2007; Wu et al. 2005). But different KR domains vary the hydroxylation pattern, generating different cyclized rings (5-methyl-1-NPA for AziB and 2-hydroxy-5-methyl-1-NPA for NcsB) (Fig. 5). An unusual off-loading mechanism was discovered in several bacterial aromatic iPKSs ─ a dedicated AT is utilized to release the polyketide products from PKSs in the biosynthesis of chlorothricin, pactamycin, avilamycin, tiacumicin B, and calicheamicin (Fig. 5) (Castillo and Perez 2008; He et al. 2009; Weitnauer et al. 2004; Weitnauer et al. 2001).
Unlike the complex multicyclic aromatic scaffolds produced by type II PKSs, aromatic polyketides produced by bacterial type I iPKSs are relatively simple and consist of a set of mono- or bicyclic aromatic products. Thus bacterial aromatic iPKSs provide a relatively simple system for understanding the catalytic mechanisms behind the synthesis of aromatic polyketides. Further understanding of these domains in bacterial aromatic iPKSs will hopefully facilitate the alterations of the functionalities on the aromatic polyketides, opening a new door to biosynthetic engineering and production of novel natural products.
Polyunsaturated fatty acid synthases
Polyunsaturated fatty acids (PUFAs) are believed beneficial to human health and nutrition. PUFAs contain multiple cis double bonds and have been identified in various marine bacteria. A gene cluster pfa from Shewanella sp. was cloned into Escherichia coli and found responsible for eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) biosynthesis (Fig. 4b) (Metz et al. 2001). Five open reading frames were subsequently found to be the minimal gene set for PUFA biosynthesis (Okuyama et al. 2007). The main PUFA iPKS can contain five to nine tandem ACP domains, in addition to the other common domains such as KS, AT, KR, and DH (Fig. 4b). After expressing the PUFA iPKSs (pfaABCDE) from S. japonica in E. coli, together with a series of in vitro and in vivo characterizations, Jiang and co-workers demonstrated that each of the predicted ACPs can be phosphopantetheinylated by the phosphopantetheinyl transferase (PPTase) in the cluster and each of the tandem ACPs is functionally equivalent for PUFA biosynthesis, but the number of ACPs controls the PUFA titer (Jiang et al. 2008). PUFA synthases from terrestrial origin, the linoleic acid producing myxobacteria, Sorangium and Aetherobacter were recently reported (Gemperlein et al. 2014). The gene organization, catalytic domain arrangement, and sequence identity of these PUFA synthases differ significantly from the marine system. 1-Acylglycerol-3-phosphate O-acyltransferase (AGPAT), a domain located at the C-terminus of the synthase, is considered to be a unique and common characteristic of terrestrial PUFA synthase. Variations also existed between the two myxobacterial pathways, such as the variable number of tandem ACPs. The significant differences among the PUFA synthases provide platform for the biosynthetic mechanism study. However, it remains unclear how the PUFA synthase complexes control the number of iteration and the level of unsaturation of the carbon chains.
Enediyne synthases
The enediynes are a family of antibiotics containing a strained ring system (9 or 10 membered), which are extremely cytotoxic (Fig. 4c) (Lam et al. 1993). The biosynthetic gene clusters for two such metabolites, the chromoprotein antibiotic C-1027 (Liu et al. 2002) and calicheamicin (Ahlert et al. 2002), have been identified. Shen and co-workers cloned the gene cluster for the 9-membered enediyne C-1027 from Streptomyces globisporus and found that an iterative type I PKS is responsible for the biosynthesis of the enediyne core (Fig. 4c) (Liu et al. 2002). The type I PKS encoded by sgcE contains six domains. Four of the domains (KS, AT, KR and DH) were in a characteristic order of known type I PKSs; one domain, unusually located at the region between AT and KR, was proposed to be an ACP; most unusually, the C-terminal domain was a PPTase domain. This arrangement is very rare but appears general to enediyne PKSs. Overall, enediyne PKSs show the greatest homology to PUFA iPKSs (Fig. 2).
The involvement of SgcE in C-1027 biosynthesis was confirmed by gene inactivation and complementation: a ΔsgcE mutant eliminated C-1027 production, and overexpression of sgcE in ΔsgcE restored C-1027 production (Liu et al. 2002). This single-module iPKS could iteratively catalyze the assembly of a linear polyunsaturated intermediate, which was further processed by a series of desaturations to furnish the two alkyne groups, then cyclized to afford the enediyne core. Indeed, a group of five to ten genes that flank the sgcE PKS gene are highly conserved and homologous either to oxidoreductases or to proteins of unknown functions. These genes seem to be only associated with enediyne biosynthesis, implying a possibly novel chemistry involved in the synthesis of the highly distinct structure of enediynes.
Another characterized enediyne is calicheamicin, which is a model for the 10-membered enediyne antibiotics (Fig. 1). The calicheamicin gene cluster was cloned from Micromonospora calichensis (Ahlert et al. 2002). The cluster contains a gene, calE8, encoding a type I PKS with the same domain organization as SgcE of C1027 (Fig. 4c). The involvement of CalE8 in calicheamicin biosynthesis was also confirmed by gene inactivation. CalE8 PKS catalyzes the biosynthesis of a polyunsaturated intermediate in an iterative process, and modifications by the associated oxidoreductases within the gene cluster then form an enediyne core intermediate (Belecki et al. 2009; Zazopoulos et al. 2003). The similarity between SgcE and CalE8 clearly suggests a common polyketide pathway for the biosynthesis of both 9- and 10-membered enediynes. SgcE and CalE8, therefore, represent a novel family of iterative type I PKSs. Townsend and co-workers further revealed insights into the biosynthesis of enediyne core structures by studying CalE8 that was heterologously expressed in E.coli (Belecki and Townsend 2012; Belecki and Townsend 2013). Their results showed that a programmed octaketide was preferentially accumulated on the enzyme and may be a key precursor to the enediyne core of calicheamicin.
Polycyclic tetramate macrolactam iPKS
Polycyclic tetramate macrolactams (PTMs) are a group of structurally distinct natural products produced by a variety of bacterial species (Fig. 4d) (Blodgett et al. 2010). A combination of small molecule chemistry, biosynthetic analysis, and genome mining in diverse bacteria led to the recognition of this group of compounds as a new class of natural products with diverse biological activities. Thus far, the most striking feature of PTM biosynthesis is that there is only one single-module PKS-NRPS hybrid gene, despite the obvious presence of two unlike polyketide moieties in the PTM skeleton (Lou et al. 2011). This suggests that this single-module PKS is responsible for the synthesis of the two polyketide moieties of PTM skeleton.
Heat–stable antifungal factor (HSAF), which has a 5,5,6-tricyclic system, was isolated from the Gram-negative bacterium Lysobacter enzymogenes (Li et al. 2006; Yu et al. 2007; Xu et al. 2015). It exhibits potent inhibitory activity against a wide range of fungi, and the studies of HSAF biosynthesis led to the recognition of the first PTM iPKS (Yu et al. 2007; Lou et al. 2011; Lou et al. 2012; Li et al. 2014). The hybrid PKS-NRPS for HSAF biosynthesis has a typical modular organization, KS-AT-DH- KR-ACP for the PKS portion and C-A-PCP-TE for the NRPS portion (Fig. 4 and 6). There is no obvious remnants of an inactive enoylreductase (ER°) domain or a methyltransferase (CMeT) domain, as seen in several iterative fungal PKS-NRPS with the similar organization, or other types of bacterial iPKSs. Although this type of iterative PKS-NRPS had been seen in fungi, it was very rare in bacteria until recent years. Phylogenetic analyses showed that PTM iPKSs are more closely related to bacterial aromatic iPKSs, rather than to fungal iPKSs (Fig. 2).
Fig. 6.
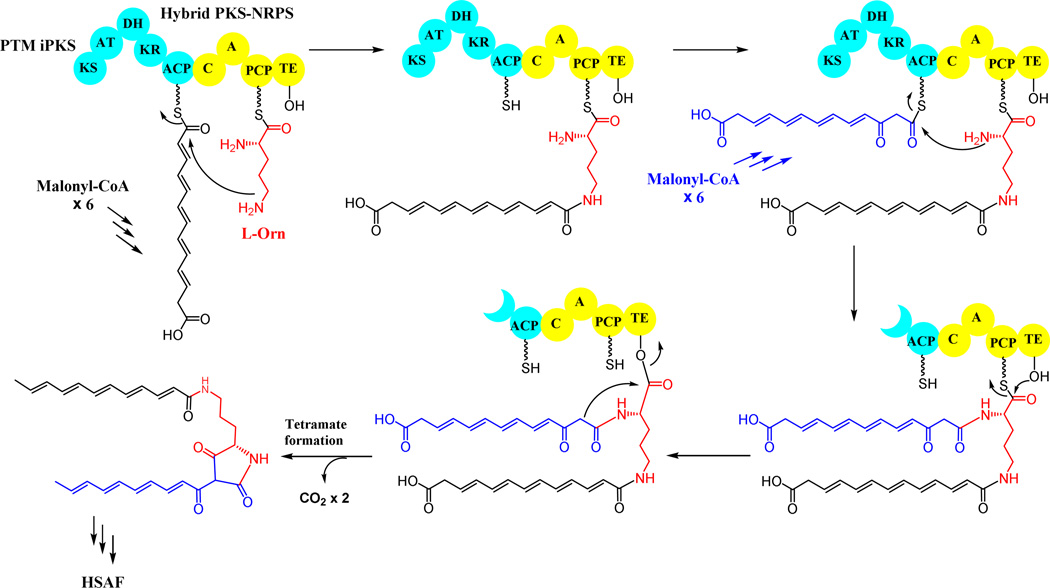
Iterative polyketide biosynthesis by PTM iPKS, a single-module, bacterial type I PKS hybridized with an NRPS module, which synthesizes two separate hexaketide chains in the biosynthesis of HSAF.
The PTM scaffold of HSAF is apparently derived from two separate hexaketide chains along with an ornithine residue. However, the biosynthetic gene cluster contains only a single PKS-NRPS and the PKS contains only a single module. This suggests that the same PKS module would act not only iteratively, but also separately, in order to construct the two different hexaketide chains and link them with the NRPS-activated ornithine to form the characteristic PTM scaffold. The HSAF biosynthetic gene cluster has been heterologously expressed in Streptomyces hosts, in which the native PKSs have been deleted, and showed that the new hosts produced HSAF analogues (Li et al. 2014). The iterative polyketide biosynthesis was also demonstrated in vitro using purified PKS and NRPS proteins. A hexaketide polyene was detected as the biosynthetic intermediate in the in vitro reactions, indicating that the single-module PKS is indeed an iterative PKS ─ it assembles two separate polyketide chains by catalyzing five cycles of chain elongation for each of the hexaketides (Fig. 6). The timing of the transfer of the second hexaketide chain is interesting, since it likely occurs immediately after the fifth cycle of polyketide chain elongation, before the newly formed β-keto group of the second hexaketide chain can be processed by the KR domain and DH domain. All of PTM natural products hitherto discovered retain this unprocessed β-keto group in the final structure (Fig. 4d). The determining factor for this precise timing is not known, but is possibly related to the tailoring redox enzymes that are proposed to cooperate with the PKS-NRPS in elaborating the PTM scaffold (Li et al. 2014).
Recently, the Gulder group reported heterologous expression of the ikarugamycin biosynthetic gene cluster in E. coli (Antosch et al. 2014), and the Zhang group reported the enzymatic mechanism for formation of the inner 5-memebered ring and demonstrated the polyketide origin of the ikarugamycin skeleton (Zhang et al. 2014). Ikarugamycin is a Streptomyces-derived PTM which has a 5,6,5-tricyclic system (Fig. 4d). Both the Gulder and Zhang groups showed that a three-gene cluster, including a PTM iPKS, is sufficient for ikarugamycin biosynthesis. In light of the huge number of uninvestigated PTM-type gene clusters in databases, studies of PTM iPKS should facilitate research into a new area of iterative bacterial type I polyketides. Improved understanding of the iterative mechanisms could be used to guide biosynthetic engineering efforts.
Partially iterative bacterial PKSs
In entirely iterative bacterial type I PKSs, the iteration events occur in the whole PKS complex. But in some bacterial type I PKSs, the iteration events can happen on only one or two modules within a multi-module assembly line. “Module stuttering” was used to describe the iterative use of only one module in a multi-module PKS, and is usually implicated when the number of modules present in a PKS is fewer than the number of condensation events required for complete biosynthesis of the corresponding polyketide (Katz 2009). Iteration can be either aberrant or programmed, and many examples of module iteration have been reported in the biosynthesis of polyketides in the recent years.
Unimodular iterations
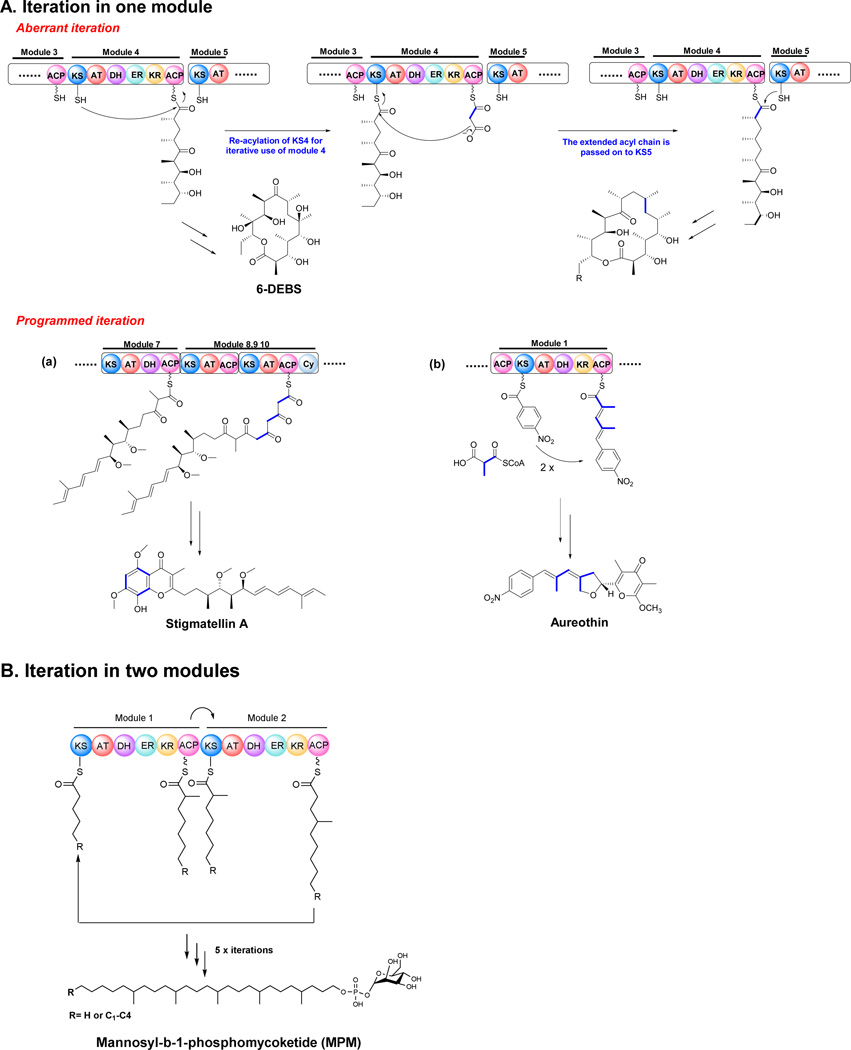
An aberrant iteration was observed in the biosynthesis of erythromycin A in S. erythraea, in which minor metabolites resulted from iterative use of one module of the PKSs (Fig. 7A) (Wilkinson et al. 2000). This was the first example of how a processive and co-linear modular PKS complex could operate in an iterative manner. This iteration is considered as aberrant because the iterative products were produced in trace amounts and the native modular PKSs generally exert a rigorous control over the chain length. Structural elucidation of the final products indicated that the iteration event occurred during the fourth chain extension (Fig. 7A). Such a homologation reaction would require the normal pentaketide product of module 4, attached to ACP4 (DEBS2), to be transferred back onto the active site cysteine residue of the KS4 domain of the same module, rather than being processed onto the downstream KS5 domain of DEBS3 (Fig. 7A). A second, module 4-catalyzed chain extension cycle would occur, producing a novel hexaketide chain attached to the ACP4 (DEBS2). This new hexaketide intermediate would be transferred to the KS5 (DEBS3) domain and the elongation process would then progress in the normal manner to give a ring-expanded octaketide product. The explanation for this aberrant iteration is that the location of the module 4 allows the iteration event to occur before the transfer of the pentaketide intermediate to the next module, but this event only happened rarely.
Fig. 7.
Iterative polyketide biosynthesis through modular iteration in bacterial type I PKS.
Similar biosynthetic iteration has been found in epothilone PKSs, which synthesize the 18-membered macrolactones (Hardt et al. 2001). Both module 5 and module 6 can be iteratively used, resulting in 6 different byproducts based on different substrate selectivity of the AT domains. The diversity of the iterative by-products indicated that the iteration events could happen to different modules within the same assembly line.
Apart from the occasional and unpredictable aberrant iteration event in some bacterial type I PKSs, most iterative PKS events are tightly controlled and apparently have developed during the evolution of the assembly lines. These programmed iterative events are found in the biosynthesis of a number of natural products. The classic examples for the programmed iterative use of a module are the type I PKSs for stigmatellin, aureothin (two iterations each), and borellidin (three iterations) (Fig. 7A).
The electron transport inhibitor stigmatellin is an aromatic compound isolated from the myxobacterium Stigmatella aurantiaca (Gaitatzis et al. 2002). By analysis of the structure and gene cluster, Muller and co-workers proposed that there must be 3 more malonyl-CoA units added to the backbone by two modules, StiH or StiJ (Fig. 7A) (Beyer et al. 1999; Gaitatzis et al. 2002). This iteration is different from the ring expansion in erythromycin biosynthesis, since stigmatellin is the only product observed in the culture broth, strongly suggesting a dedicated and programmed iterative processing in this PKS assembly. Either StiH or StiJ could be used iteratively, while the other modules worked classically. It was also possible that this iteration was actually from an in trans module outside the gene cluster. To exclude this possibility, the authors inactivated every other PKS-containing gene cluster in the genome of S. aurantiaca. However, none of these mutations eliminated the production of stigmatellin, showing that its biosynthesis is solely the responsibility of the sti locus, apparently necessitating iterative use of the associated PKS.
The biosynthesis of lankacidin is a very unusual example of the iterative use of a PKS module. A module, consisting of KS-DH-CM-ACP1-ACP1-KS-AT spread over four proteins, was proposed to be iteratively used for the first five condensations (Mochizuki et al. 2003) . The presence of repeated, virtually identical, ACP domains is also an interesting feature, suggesting that this module may be involved in iterative use, with twin ACP domains having different uses.
The aureothin PKS also contains a programmed iteratively-used module (Fig. 7A) (He and Hertweck 2003; He and Hertweck 2005; Traitcheva et al. 2007). The iteration module, AurA, not only works iteratively, but also selects the p-nitrobenzoate starter unit, revealing that the interplay of multiple components is essential to control the exact number of elongation cycles (Busch et al. 2013). Hertweck and co-workers proposed that the iteration happened by retrotransfer of the biosynthetic intermediate from one PKS subunit to the opposite PKS subunit of the synthase complex (Busch et al. 2012). They confirmed that KS1 was in charge of priming the PKS, thus leaving a vacant position for the retrotransfer of the diketide. KS2 functions as a gatekeeper that controls the chain length of polyketide. The substitution of module 1 of AurA to the avemectin module 1 resulted in abolition of iteration, indicating that iteration is completely due to aureothin module 1. The deletion of the TE domain enabled the authors to track the intermediates produced by aur PKS (Busch et al. 2013). The biosynthetic logic found in aureothin PKS was used to morph an artificial pathway, which generates a complex polyketide that was initially isolated from a different bacterium (Sugimoto et al. 2014). This is the first example of engineering modular PKS to generate known bioactive compounds. The synthase of neoaureothin, a homolog of aureothin, also employs the module 1 iteratively, but allows additional chain elongation. The resulting derivatives of the truncated nor PKS variants suggested the iterative modules might compensate for the loss of a module, while downstream modules serve as the gatekeepers to control the chain length (Sugimoto et al. 2015). Iterative PKS programming was also found in the biosynthesis of etnangien (Menche et al. 2008) and crocacin (Muller et al. 2014). The investigation of programmed module iterations may lead to new diversity of polyketide compounds through combinatorial biosynthesis.
Bimodular iterations
PKS module iteration was initially thought to involve only one module. However, a novel bimodular iteration was revealed in 2008 (Chopra et al. 2008). A protein involved in the biosynthesis of a phosphoglycolipid called mannosyl-β-1-phosphomycoketide (MPM) was proposed to contain two complete sets of modules and to synthesize mycoketide by five alternating condensations of methylmalonyl and malonyl units, using an iterative mode of PKS catalysis (instead of terpene synthase catalysis) (Fig. 7B). The chemical structure of mycoketide contains branching at every alternate ketide unit, suggesting an alternative use of two modules wherein one module would condense a branched C3 ketide unit and the next would add a C2 unit. Site mutation and radiolabeled experiments confirmed that the AT1 prefers methylmalonyl-CoA while the AT2 recruits malonyl-CoA. On the basis of biochemical, computational, mutagenic, analytical ultracentrifugation and atomic force microscopic studies, it was proposed that the PKS12 protein could be organized as a large supramolecular assembly mediated through specific interactions between the C- and N-terminus linkers. This interaction enables the intermolecular acyl chain transfer between two proteins. PKS12 protein thus forms a modular assembly to perform repetitive condensations analogous to iterative proteins (Chopra et al. 2008). This novel intermolecular iterative biosynthetic mechanism provides new perspective on the understanding of polyketide biosynthetic machinery and also suggests new ways to engineer polyketide metabolites.
Iterative mechanisms
Khosla and co-workers proposed a ratchet mechanism for the naturally noniterative PKS assembly line, that is, the first helix of ACP domain is preferentially recognized by the KS of next module instead of the one in its own module (Kapur et al. 2012). The KS-ACP recognition and interaction allow unidirectional translocation of the growing polyketide chain. Upon this theory, an intrinsically noniterative PKS could be engineered to be iterative by rational swapping the first ACP helix. Indeed, a noniterative DEBS module has been engineered to catalyze two rounds of chain elongation by replacing ACP helix I from module 3 to ACP helix I from module 2 (Kapur et al. 2012).
Although many examples of bacterial type I iPKSs have been reported, the mechanism that controls the iteration processes in these iPKSs has not been clearly elucidated (Busch and Hertweck 2009). A probable mechanism could be that iteration is an intrinsic feature of the module like FAS and fungal iPKSs. Alternatively, a “force” coming from the downstream, for example, the KS domain which only accepts an intermediate with a particular chain length, could encourage or restrict the upstream module to iterate, passing the nascent polyketide back upstream for another round of elongation. A combination of both is possible in some of the reported cases (Busch et al. 2013). The number or “freedom” of the iteration is influenced by the intrinsic property of the iterative module itself, but the size of the final product is determined by the selectivity of the downstream gatekeepers (Sugimoto et al. 2015).
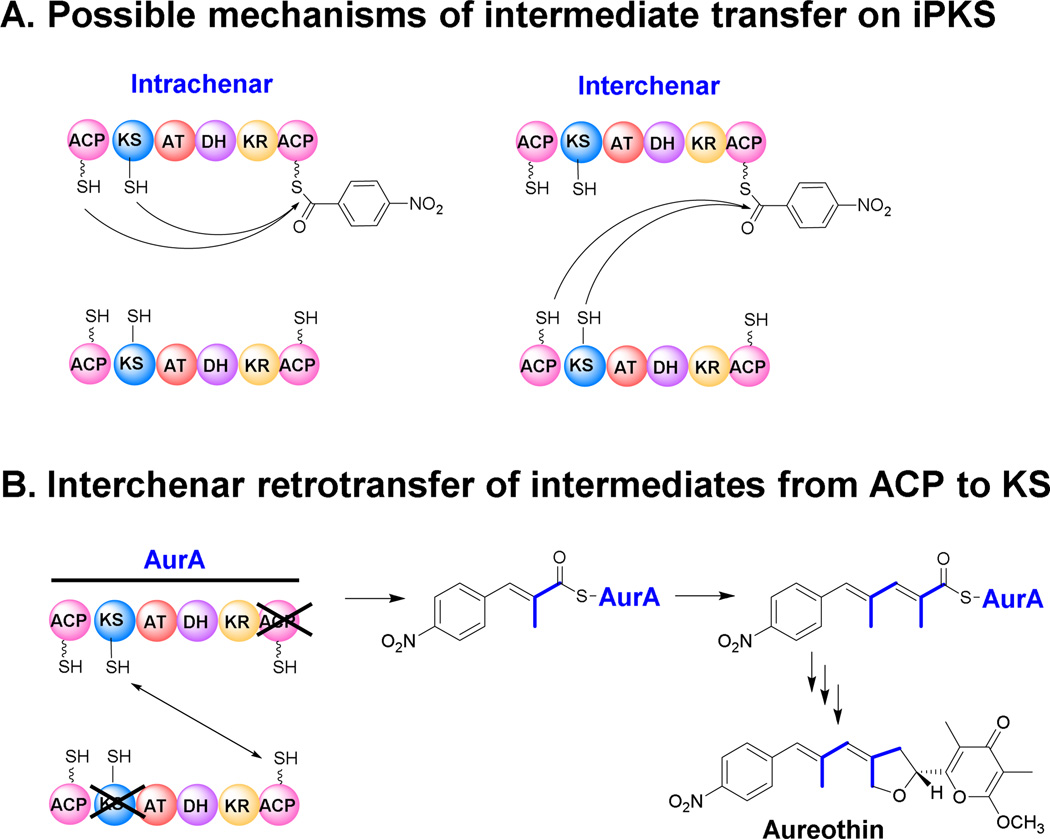
Despite the lack of experimental evidence in general, the process of iteration in bacterial type I iPKSs could be mirrored on what has been described for fungal iPKSs: (1) KS domain catalyzes the Claisen condensation reaction between the starter and extender units, driven by decarboxylation of the extender unit; (2) the intermediate is passed back to the KS domain to be extended by another ketide unit; (3) the finished polyketide chain is released from the enzyme only once the polyketide has reached its predetermined length, which is unique for each iPKS. In principle, the iteration might happen under the following scenarios: the intermediate is transferred from the ACP domain of one PKS chain to (a) ACP domain on the same PKS chain (‘intrachenar’), (b) ACP domain on the opposite PKS chain (‘interchenar’), (c) KS domain upstream on the same PKS chain (‘intrachenar’), or (d) KS domain upstream on the opposite chain (‘interchenar’) (Fig. 8). Through targeted domain mutagenesis, cross-complementation experiments, and metabolic profiling, Hertweck and co-workers proposed an “interchenar retrotransfer” model for the iPKS involved in aureothin biosynthesis (Busch et al. 2012). They revealed that the N-terminus ACP is not involved in the iteration process, ruling out an ACP-ACP shuttle (Fig. 8). This is also consistent with the fact that most bacterial iPKSs don’t have an N-terminal ACP. Furthermore, an aurA (ΔKS, ΔACP) and aurA (ΔAT) heterodimer proved to be nonfunctional, whereas aureothin production was restored in a ΔaurA mutant complemented with aurA (ΔKS) - aurA (ΔACP). This finding supports that the most likely scenario is (d), a retrotransfer of the biosynthetic intermediate from the ACP onto the KS domain located on the opposite polypeptide strand. Finally, evidence also suggested that iteration events in a modular polyketide assembly line are controlled by multiple factors (Busch et al. 2013). The interplay of multiple components is essential not only to the exact number of iteration cycles, but also to an unobstructed metabolite flux in the assembly line. The kinetics can be an important factor in this process. For example in aureothin PKS, the KS1 of AurA is exclusively in charge of priming the PKS and the retrotransfer of the diketide, and the KS2 of AurB is a gatekeeper only accepting an intermediate with a right size. For a programmed iteration to take place, KS1 must control the rates of priming and retrotransfer of the diketide. When a designated loading module of the avermectin PKS was placed in front of AurA, the iteration is completely lost in the engineered PKS (Busch et al. 2013).
Fig. 8.
Possible mechanisms of iteration in a single-module type I iPKS, intrachenar transfer versus interchenar transfer (A) and the experimentally supported interchenal retrotransfer in aureothin biosynthesis (B).
Final remarks
PKSs are generally classified into three types according to their organizations. With the increasing number of PKSs falling into the category of “non-canonical” PKSs, the boundaries between the different types are no longer so well defined. Non-canonical type I PKSs expand the diversity of natural product biosynthesis, making engineering and rational design in synthetic biology even more complicated. Iteration is likely the most unpredictable event that occurs in polyketide biosynthesis. The iterations can happen within one or two modules of a giant multi-module assembly line, and iterations can occur either once or multiple times. In the case of AurA, the iterative module can be repetitively used for four times until the length of the intermediate is selected by the next KS gatekeeper. For entirely iterative PKSs, such as those for bacterial aromatic polyketides, PTM, PUFA, and enediynes, these modules have the potential to provide the entire core structures of the target compounds, which could also be further engineered to produce libraries of complex polyketides. Programmed iteration may have the potential to be engineered to produce polyketides with diverse lengths and modifications.
However, the realization of engineered biosynthesis relies on the understanding of the molecular basis behind iterations in bacterial type I PKSs. Although several groups have begun to investigate the mechanisms of iteration and timing of retrotransfer, more work has to be done to understand these processes and to translate the knowledge into polyketide biosynthetic engineering. At the same time, these iterative events continue to reveal that Nature recruits “unexpected” mechanisms to synthesize novel natural products. With the progress in PKS structural biology and in depth biochemical studies, we anticipate that new insights into the molecular mechanisms by which bacterial modular PKSs control iterations will continue to emerge.
Acknowledgements
Funding: This study was supported in part by the NIH (R01AI097260), NSFC (31329005), and a University of Nebraska-Lincoln Redox Biology Center pilot grant.
Footnotes
Ethical Statement
Conflict of Interest: Haotong Chen declares that she has no conflict of interest; Liangcheng Du declares that he has no conflict of interest.
Ethical approval: This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang KX, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- Antosch J, Schaefers F, Gulder TA. Heterologous reconstitution of ikarugamycin biosynthesis in E. coli . Angew Chem Int Edit. 2014;53:3011–3014. doi: 10.1002/anie.201310641. [DOI] [PubMed] [Google Scholar]
- Belecki K, Crawford JM, Townsend CA. Production of octaketide polyenes by the calicheamicin polyketide synthase CalE8: Implications for the biosynthesis of enediyne core structures. J Am Chem Soc. 2009;131:12564–12566. doi: 10.1021/ja904391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belecki K, Townsend CA. Environmental control of the calicheamicin polyketide synthase leads to detection of a programmed octaketide and a proposal for enediyne biosynthesis. Angew Chem Int Edit. 2012;51:11316–11319. doi: 10.1002/anie.201206462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belecki K, Townsend CA. Biochemical determination of enzyme-bound metabolites: preferential accumulation of a programmed octaketide on the enediyne polyketide synthase CalE8. J Am Chem Soc. 2013;135:14339–14348. doi: 10.1021/ja406697t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer S, Kunze B, Silakowski B, Muller R. Metabolic diversity in myxobacteria: identification of the myxalamid and the stigmatellin biosynthetic gene cluster of Stigmatella aurantiaca Sg a15 and a combined polyketide-(poly)peptide gene cluster from the epothilone producing strain Sorangium cellulosum So ce90. Biochim Biophys Acta. 1999;1445:185–195. doi: 10.1016/s0167-4781(99)00041-x. [DOI] [PubMed] [Google Scholar]
- Blodgett JAV, Oh DC, Cao SG, Currie CR, Kolter R, Clardy J. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc Natl Acad Sci USA. 2010;107:11692–11697. doi: 10.1073/pnas.1001513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider T, Heim JB, Heine D, Winkler R, Busch B, Kusebauch B, Stehle T, Zocher G, Hertweck C. Vinylogous chain branching catalysed by a dedicated polyketide synthase module. Nature. 2013;502:124–128. doi: 10.1038/nature12588. [DOI] [PubMed] [Google Scholar]
- Broadhurst RW, Nietlispach D, Wheatcroft MP, Leadlay PF, Weissman KJ. The structure of docking domains in modular polyketide synthases. Chem Biol. 2003;10:723–731. doi: 10.1016/s1074-5521(03)00156-x. [DOI] [PubMed] [Google Scholar]
- Busch B, Hertweck C. Evolution of metabolic diversity in polyketide-derived pyrones: Using the non-colinear aureothin assembly line as a model system. Phytochem. 2009;70:1833–1840. doi: 10.1016/j.phytochem.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Busch B, Ueberschaar N, Behnken S, Sugimoto Y, Werneburg M, Traitcheva N, He J, Hertweck C. Multifactorial control of iteration events in a modular polyketide assembly line. Angew Chem Int Edit. 2013;52:5285–5289. doi: 10.1002/anie.201301322. [DOI] [PubMed] [Google Scholar]
- Busch B, Ueberschaar N, Sugimoto Y, Hertweck C. Interchenar retrotransfer of aureothin intermediates in an iterative polyketide synthase module. J Am Chem Soc. 2012;134:12382–12385. doi: 10.1021/ja304454r. [DOI] [PubMed] [Google Scholar]
- Castillo YP, Perez MA. Bacterial beta-ketoacyl-acyl carrier protein synthase III (FabH): an attractive target for the design of new broad-spectrum antimicrobial agents. Mini Rev Med Chem. 2008;8:36–45. doi: 10.2174/138955708783331559. [DOI] [PubMed] [Google Scholar]
- Challis GL. Genome mining for novel natural product discovery. J Med Chem. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Zhao J, Liu W, Gao JF, Tao LM, Pan HX, Tang GL. Identification of phoslactomycin biosynthetic gene clusters from Streptomyces platensis SAM-0654 and characterization of PnR1 and PnR2 as positive transcriptional regulators. Gene. 2012;509:195–200. doi: 10.1016/j.gene.2012.08.030. [DOI] [PubMed] [Google Scholar]
- Cheng YQ, Tang GL, Shen B. Identification and localization of the gene cluster encoding biosynthesis of the antitumor macrolactam leinamycin in Streptomyces atroolivaceus S-140. J Bacteriol. 2002;184:7013–7024. doi: 10.1128/JB.184.24.7013-7024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YQ, Tang GL, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Natl Acad Sci USA. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra T, Banerjee S, Gupta S, Yadav G, Anand S, Surolia A, Roy RP, Mohanty D, Gokhale RS. Novel intermolecular iterative mechanism for biosynthesis of mycoketide synthase by a bimodular polyketide synthase. PLoS Biol. 2008;6:1584–1598. doi: 10.1371/journal.pbio.0060163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea . Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Townsend CA. New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8:879–889. doi: 10.1038/nrmicro2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum M, Peintner I, Linnenbrink A, Frerich A, Weber M, Paululat T, Bechthold A. Organization of the biosynthetic gene cluster and tailoring enzymes in the biosynthesis of the tetracyclic quinone glycoside antibiotic polyketomycin. ChemBioChem. 2009;10:1073–1083. doi: 10.1002/cbic.200800823. [DOI] [PubMed] [Google Scholar]
- Ding W, Deng W, Tang MC, Zhang Q, Tang GL, Bi YR, Liu W. Biosynthesis of 3-methoxy-5-methyl naphthoic acid and its incorporation into the antitumor antibiotic azinomycin B. Mol Biosyst. 2010;6:1071–1081. doi: 10.1039/b926358f. [DOI] [PubMed] [Google Scholar]
- Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- El-Sayed AK, Hothersall J, Cooper SM, Stephens E, Simpson TJ, Thomas CM. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem Biol. 2003;10:419–430. doi: 10.1016/s1074-5521(03)00091-7. [DOI] [PubMed] [Google Scholar]
- Fisch KM. Biosynthesis of natural products by microbial iterative hybrid PKS-NRPS. Rsc Adv. 2013;3:18228–18247. [Google Scholar]
- Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Fujii I. Functional analysis of fungal polyketide biosynthesis genes. J Antibiot. 2010;63:207–218. doi: 10.1038/ja.2010.17. [DOI] [PubMed] [Google Scholar]
- Gaisser S, Trefzer A, Stockert S, Kirschning A, Bechthold A. Cloning of an avilamycin biosynthetic gene cluster from Streptomyces viridochromogenes Tu57. J Bacteriol. 1997;179:6271–6278. doi: 10.1128/jb.179.20.6271-6278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitatzis N, Silakowski B, Kunze B, Nordsiek G, Blocker H, Hofle G, Muller R. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J Biol Chem. 2002;277:13082–13090. doi: 10.1074/jbc.M111738200. [DOI] [PubMed] [Google Scholar]
- Gay DC, Gay G, Axelrod AJ, Jenner M, Kohlhaas C, Kampa A, Oldham NJ, Piel J, Keatinge-Clay AT. A close look at a ketosynthase from a trans-acyltransferase modular polyketide synthase. Structure. 2014;22:444–451. doi: 10.1016/j.str.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemperlein K, Rachid S, Garcia RO, Wenzel SC, Muller R. Polyunsaturated fatty acid biosynthesis in myxobacteria: different PUFA synthases and their product diversity. Chem Sci. 2014;5:1733–1741. [Google Scholar]
- Hardt IH, Steinmetz H, Gerth K, Sasse F, Reichenbach H, Hofle G. New natural epothilones from Sorangium cellulosum, strains So ce90/B2 and So ce90/D13: Isolation, structure elucidation, and SAR studies. J Nat Prod. 2001;64:847–856. doi: 10.1021/np000629f. [DOI] [PubMed] [Google Scholar]
- He J, Hertweck C. Iteration as programmed event during polyketide assembly; molecular analysis of the aureothin biosynthesis gene cluster. Chem Biol. 2003;10:1225–1232. doi: 10.1016/j.chembiol.2003.11.009. [DOI] [PubMed] [Google Scholar]
- He J, Hertweck C. Functional analysis of the aureothin iterative type I polyketide synthase. ChemBioChem. 2005;6:908–912. doi: 10.1002/cbic.200400333. [DOI] [PubMed] [Google Scholar]
- He Q, Jia X, Tang M, Tian Z, Tang G, Liu W. Dissection of two acyl-transfer reactions centered on acyl-S-carrier protein intermediates for incorporating 5-chloro-6-methyl-O-methylsalicyclic acid into chlorothricin. ChemBioChem. 2009;10:813–819. doi: 10.1002/cbic.200800714. [DOI] [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Edit. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- Hertweck C. Decoding and reprogramming complex polyketide assembly lines: prospects for synthetic biology. Trends Biochem Sci. 2015;40:189–199. doi: 10.1016/j.tibs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- Ito T, Roongsawang N, Shirasaka N, Lu WL, Flatt PM, Kasanah N, Miranda C, Mahmud T. Deciphering pactamycin biosynthesis and engineered production of new pactamycin analogues. ChemBioChem. 2009;10:2253–2265. doi: 10.1002/cbic.200900339. [DOI] [PubMed] [Google Scholar]
- Jenke-Kodama H, Sandmann A, Muller R, Dittmann E. Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol. 2005;22:2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- Jia XY, Tian ZH, Shao L, Qu XD, Zhao QF, Tang J, Tang GL, Liu W. Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem Biol. 2006;13:575–585. doi: 10.1016/j.chembiol.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Jiang H, Zirkle R, Metz JG, Braun L, Richter L, Van Lanen SG, Shen B. The role of tandem acyl carrier protein domains in polyunsaturated fatty acid biosynthesis. J Am Chem Soc. 2008;130:6336–6337. doi: 10.1021/ja801911t. [DOI] [PubMed] [Google Scholar]
- Julien B, Shah S, Ziermann R, Goldman R, Katz L, Khosla C. Isolation and characterization of the epothilone biosynthetic gene cluster from Sorangium cellulosum . Gene. 2000;249:153–160. doi: 10.1016/s0378-1119(00)00149-9. [DOI] [PubMed] [Google Scholar]
- Kapur S, Lowry B, Yuzawa S, Kenthirapalan S, Chen AY, Cane DE, Khosla C. Reprogramming a module of the 6-deoxyerythronolide B synthase for iterative chain elongation. Proc Natl Acad Sci USA. 2012;109:4110–4115. doi: 10.1073/pnas.1118734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L. The DEBS paradigm for type I modular polyketide synthases and beyond. Meth Enzymol. 2009;459:113–142. doi: 10.1016/S0076-6879(09)04606-0. [DOI] [PubMed] [Google Scholar]
- Kaulmann U, Hertweck C. Biosynthesis of polyunsaturated fatty acids by polyketide synthases. Angew Chem Int Edit. 2002;41:1866–1869. doi: 10.1002/1521-3773(20020603)41:11<1866::aid-anie1866>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- Khosla C. Natural product biosynthesis: A new interface between enzymology and medicine. J Org Chem. 2000;65:8127–8133. doi: 10.1021/jo000849y. [DOI] [PubMed] [Google Scholar]
- Khosla C, Kapur S, Cane DE. Revisiting the modularity of modular polyketide synthases. Curr Opin Chem Biol. 2009;13:135–143. doi: 10.1016/j.cbpa.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KS, Veitch JA, Golik J, Krishnan B, Klohr SE, Volk KJ, Forenza S, Doyle TW. Biosynthesis of esperamicin A1, an enediyne antitumor antibiotic. J Am Chem Soc. 1993;115:12340–12345. [Google Scholar]
- Li S, Du L, Yuen G, Harris SD. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans . Mol Biol Cell. 2006;17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen H, Ding Y, Xie Y, Wang H, Cerny RL, Shen Y, Du L. Iterative assembly of two separate polyketide chains by the same single-module bacterial polyketide synthase in the biosynthesis of HSAF. Angew Chem Int Edit. 2014;53:7524–7530. doi: 10.1002/anie.201403500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu X, Seipke RF, Zhang W. Biosynthesis of antimycins with a reconstituted 3-formamidosalicylate pharmacophore in Escherichia coli . ACS Synth Biol. 2015a;4:559–565. doi: 10.1021/sb5003136. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang Z, Shao CL, Wang JL, Bai H, Wang CY. Bioinformatical analysis of the sequences, structures and functions of fungal polyketide synthase product template domains. Scientific Reports. 2015b;5:10463. doi: 10.1038/srep10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes . J Am Chem Soc. 2011;133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou L, Chen H, Cerny RL, Li Y, Shen Y, Du L. Unusual activities of the thioesterase domain for the biosynthesis of the polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes C3. Biochemistry. 2012;51:4–6. doi: 10.1021/bi2015025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menche D, Arikan F, Perlova O, Horstmann N, Ahlbrecht W, Wenzel SC, Jansen R, Irschik H, Muller R. Stereochemical determination and complex biosynthetic assembly of etnangien, a highly potent RNA polymerase inhibitor from the myxobacterium Sorangium cellulosum . J Am Chem Soc. 2008;130:14234–14243. doi: 10.1021/ja804194c. [DOI] [PubMed] [Google Scholar]
- Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domergue F, Yamada A, Yazawa K, Knauf V, Browse J. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 2001;293:290–293. doi: 10.1126/science.1059593. [DOI] [PubMed] [Google Scholar]
- Mochizuki S, Hiratsu K, Suwa M, Ishii T, Sugino F, Yamada K, Kinashi H. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol Microbiol. 2003;48:1501–1510. doi: 10.1046/j.1365-2958.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- Molnar I, Schupp T, Ono M, Zirkle R, Milnamow M, Nowak-Thompson B, Engel N, Toupet C, Stratmann A, Cyr DD, Gorlach J, Mayo JM, Hu A, Goff S, Schmid J, Ligon JM. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem Biol. 2000;7:97–109. doi: 10.1016/s1074-5521(00)00075-2. [DOI] [PubMed] [Google Scholar]
- Moore BS, Hopke JN. Discovery of a new bacterial polyketide biosynthetic pathway. ChemBioChem. 2001;2:35–38. doi: 10.1002/1439-7633(20010105)2:1<35::AID-CBIC35>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Moss SJ, Martin CJ, Wilkinson B. Loss of co-linearity by modular polyketide synthases: a mechanism for the evolution of chemical diversity. Nat Prod Rep. 2004;21:575–593. doi: 10.1039/b315020h. [DOI] [PubMed] [Google Scholar]
- Muller R. Don’t classify polyketide synthases. Chem Biol. 2004;11:4–6. doi: 10.1016/j.chembiol.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Muller S, Rachid S, Hoffmann T, Surup F, Volz C, Zaburannyi N, Muller R. Biosynthesis of crocacin involves an unusual hydrolytic release domain showing similarity to condensation domains. Chem Biol. 2014;21:855–865. doi: 10.1016/j.chembiol.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Musiol EM, Hartner T, Kulik A, Moldenhauer J, Piel J, Wohlleben W, Weber T. Supramolecular templating in kirromycin biosynthesis: the acyltransferase KirCII loads ethylmalonyl-CoA extender onto a specific ACP of the trans-AT PKS. Chem Biol. 2011;18:438–444. doi: 10.1016/j.chembiol.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Musiol EM, Weber T. Discrete acyltransferases involved in polyketide biosynthesis. MedChemComm. 2012;3:871–886. [Google Scholar]
- Okuyama H, Orikasa Y, Nishida T, Watanabe K, Morita N. Bacterial genes responsible for the biosynthesis of eicosapentaenoic and docosahexaenoic acids and their heterologous expression. Appl Environ Microbiol. 2007;73:665–670. doi: 10.1128/AEM.02270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan N, Alhamadsheh MM, Reynolds KA. cis-Delta(2,3)-double bond of phoslactomycins is generated by a post-PKS tailoring enzyme. J Am Chem Soc. 2008;130:12236–12237. doi: 10.1021/ja8044162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- Shen B, Cheng YQ, Christenson SD, Jiangi H, Ju JH, Kwon HJ, Lim SK, Liu W, Nonaka K, Seo JW, Smith WC, Standage S, Tang GL, Van Lanen S, Zhang J. Polyketide biosynthesis beyond the Type I, II, and III polyketide synthase paradigms: a progress report. Acs Sym Ser. 2007;955:154–166. [Google Scholar]
- Shen B, Thorson JS. Expanding nature’s chemical repertoire through metabolic engineering and biocatalysis. Curr Opin Chem Biol. 2012;16:99–100. doi: 10.1016/j.cbpa.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Sherman DH. The Lego-ization of polyketide biosynthesis. Nat Biotechnol. 2005;23:1083–1084. doi: 10.1038/nbt0905-1083. [DOI] [PubMed] [Google Scholar]
- Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Ding L, Ishida K, Hertweck C. Rational design of modular polyketide synthases: morphing the aureothin pathway into a luteoreticulin assembly line. Angew Chem Int Edit. 2014;53:1560–1564. doi: 10.1002/anie.201308176. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Ishida K, Traitcheva N, Busch B, Dahse HM, Hertweck C. Freedom and constraint in engineered noncolinear polyketide assembly lines. Chem Biol. 2015;22:229–240. doi: 10.1016/j.chembiol.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Sun H, Ho C, Ding F, Soehano I, Liu X, Liang Z. Synthesis of (R)-mellein by a partially reducing iterative polyketide synthase. J Am Chem Soc. 2012;134:11924–11927. doi: 10.1021/ja304905e. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Toyoda A, Sekiyama Y, Takagi H, Nogawa T, Uramoto M, Suzuki R, Koshino H, Kumano T, Panthee S, Dairi T, Ishikawa J, Ikeda H, Sakaki Y, Osada H. Reveromycin A biosynthesis uses RevG and RevJ for stereospecific spiroacetal formation. Nat Chem Biol. 2011;7:461–468. doi: 10.1038/nchembio.583. [DOI] [PubMed] [Google Scholar]
- Traitcheva N, Jenke-Kodama H, He J, Dittmann E, Hertweck C. Non-colinear polyketide biosynthesis in the aureothin and neoaureothin pathways: An evolutionary perspective. ChemBioChem. 2007;8:1841–1849. doi: 10.1002/cbic.200700309. [DOI] [PubMed] [Google Scholar]
- Van Lanen SG, Lin SJ, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027 involves a new branching point in chorismate metabolism. Proc Natl Acad Sci USA. 2008;105:494–499. doi: 10.1073/pnas.0708750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B. Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J Am Chem Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K. Exploring the biosynthesis of natural products and their inherent suitability for the rational design of desirable compounds through genetic engineering. Biosci Biotech Bioch. 2008;72:2491–2506. doi: 10.1271/bbb.80323. [DOI] [PubMed] [Google Scholar]
- Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- Weitnauer G, Hauser G, Hofmann C, Linder U, Boll R, Pelz K, Glaser SJ, Bechthold A. Novel avilamycin derivatives with improved polarity generated by targeted gene disruption. Chem Biol. 2004;11:1403–1411. doi: 10.1016/j.chembiol.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Weitnauer G, Muhlenweg A, Trefzer A, Hoffmeister D, Sussmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Biosynthesis of the orthosomycin antibiotic avilamycin A: deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tu57 and production of new antibiotics. Chem Biol. 2001;8:569–581. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Foster G, Rudd BAM, Taylor NL, Blackaby AP, Sidebottom PJ, Cooper DJ, Dawson MJ, Buss AD, Gaisser S, Bohm IU, Rowe CJ, Cortes J, Leadlay PF, Staunton J. Novel octaketide macrolides related to 6-deoxyerythronolide B provide evidence for iterative operation of the erythromycin polyketide synthase. Chem Biol. 2000;7:111–117. doi: 10.1016/s1074-5521(00)00076-4. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Kendrew SG, Sheridan RM, Leadlay PF. Biosynthetic engineering of polyketide synthases. Expert Opin Ther Pat. 2003;13:1579–1606. [Google Scholar]
- Williams GJ. Engineering polyketide synthases and nonribosomal peptide synthetases. Curr Opin Struc Biol. 2013;23:603–612. doi: 10.1016/j.sbi.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr Opin Chem Biol. 2011;15:22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Winter JM, Tang Y. Synthetic biological approaches to natural product biosynthesis. Curr Opin Biotech. 2012;23:736–743. doi: 10.1016/j.copbio.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zaleski TJ, Valenzano C, Khosla C, Cane DE. Polyketide double bond biosynthesis. Mechanistic analysis of the dehydratase-containing module 2 of the picromycin/methymycin polyketide synthase. J Am Chem Soc. 2005;127:17393–17404. doi: 10.1021/ja055672+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Li S, Niu S, Ma L, Zhang G, Zhang H, Zhang G, Ju J, Zhang C. Characterization of tiacumicin B biosynthetic gene cluster affording diversified tiacumicin analogues and revealing a tailoring dihalogenase. J Am Chem Soc. 2011;133:1092–1105. doi: 10.1021/ja109445q. [DOI] [PubMed] [Google Scholar]
- Xu L, Wu P, Wright SJ, Du L, Wei X. Bioactive polycyclic tetramate macrolactams from Lysobacter enzymogenes and their absolute configurations by theoretical ECD calculations. J Nat Prod. 2015;78:1841–1847. doi: 10.1021/acs.jnatprod.5b00099. [DOI] [PubMed] [Google Scholar]
- Xu W, Qiao KJ, Tang Y. Structural analysis of protein-protein interactions in type I polyketide synthases. Crit Rev Biochem Mol Biol. 2013;48:98–122. doi: 10.3109/10409238.2012.745476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala AO, Cacho RA, Tang Y. Protein engineering towards natural product synthesis and diversification. J Ind Microbiol Biot. 2012;39:227–241. doi: 10.1007/s10295-011-1044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazopoulos E, Huang KX, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- Zhang G, Zhang W, Zhang Q, Shi T, Ma L, Zhu Y, Li S, Zhang H, Zhao Y, Shi R, Zhang C. Mechanistic insights into polycycle formation by reductive cyclization in ikarugamycin biosynthesis. Angew Chem Int Edit. 2014;53:4840–4844. doi: 10.1002/anie.201402078. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Pang B, Ding W, Liu W. Aromatic polyketides produced by bacterial iterative type I polyketide synthases. ACS Catal. 2013;3:1439–1447. [Google Scholar]
- Zhang W, Tang Y. In vitro analysis of type II polyketide synthase. Methods in Enzymology: Microbial Natural Product Biosynthesis. 2009;459:367–393. doi: 10.1016/S0076-6879(09)04616-3. [DOI] [PubMed] [Google Scholar]
- Zhao QF, He QL, Ding W, Tang MC, Kang QJ, Yu Y, Deng W, Zhang Q, Fang J, Tang GL, Liu W. Characterization of the azinomycin B biosynthetic gene cluster revealing a different iterative type I polyketide synthase for naphthoate biosynthesis. Chem Biol. 2008;15:693–705. doi: 10.1016/j.chembiol.2008.05.021. [DOI] [PubMed] [Google Scholar]
- Zou Y, Yin H, Kong D, Deng Z, Lin S. A trans-acting ketoreductase in biosynthesis of a symmetric polyketide dimer SIA7248. ChemBioChem. 2013;14:679–683. doi: 10.1002/cbic.201300068. [DOI] [PubMed] [Google Scholar]