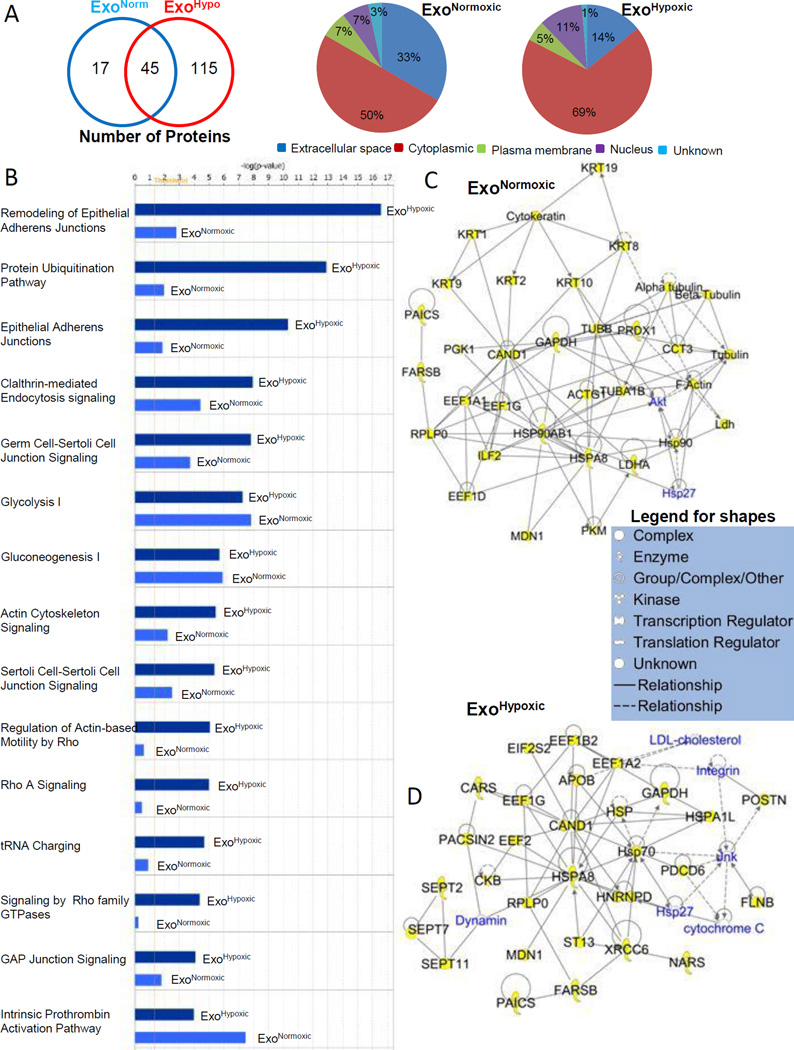
Figure 5. Characterization of ExoNormoxic and ExoHypoxic proteins by mass spectrometry.
(A) Total proteins (both distinct and overlapping) present in ExoNormoxic and ExoHypoxic are presented in a Venn diagram (Left Panel). Proteins were characterized using Ingenuity IPA software, and proteins subcellular (or extracellular) localization is presented in pie diagrams (Middle and Right Panel). (B) ExoNormoxic and ExoHypoxic total protein data were compared using Ingenuity IPA software. Comparisons of the Top 15 canonical pathways are shown. Scores (total number of proteins in the sample versus total number of known proteins of that pathway) are plotted as –log value, which is derived from a p-value and indicates the likelihood of the Focus Proteins in a network being found together due to random chance. Threshold value is set at 1.25. (C–D) ExoNormoxic and ExoHypoxic proteins clustered within the Top Networks/Associated Functions as derived from IPA algorithms are shown as members of the “interactomes”. Protein shapes are indicative of function and that legend is shown.