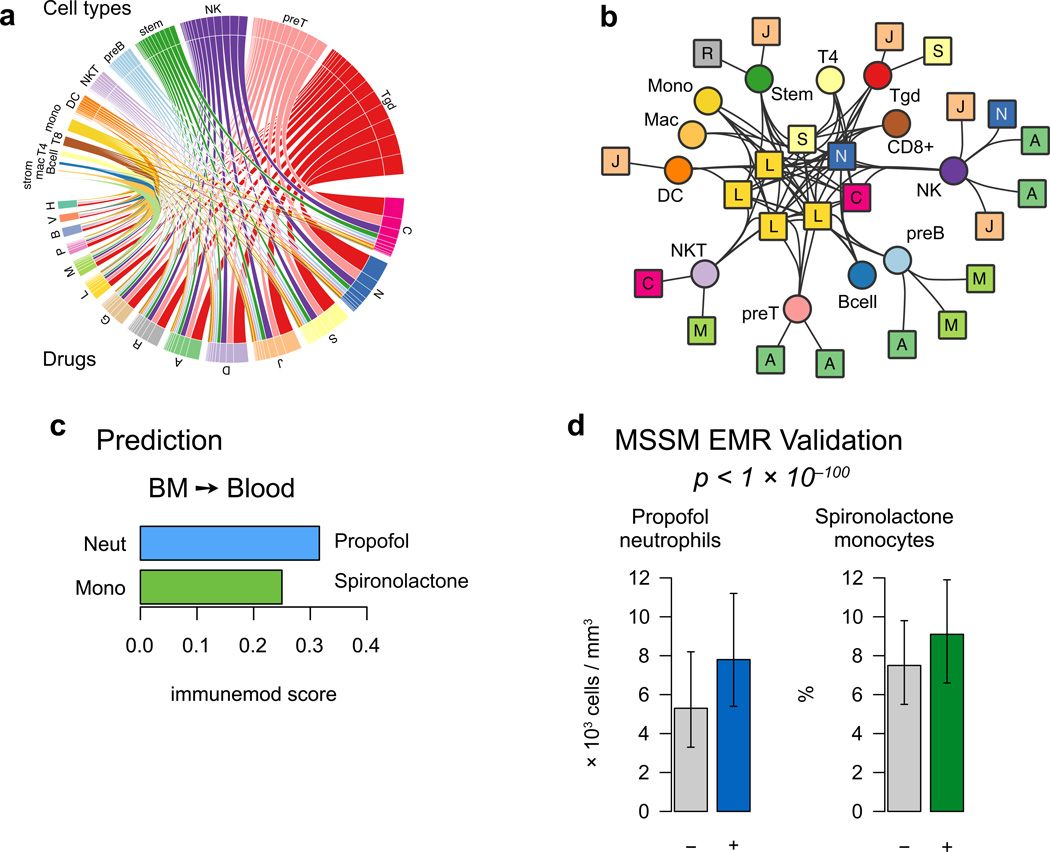
Figure 3. Predicting immune cell influence from the IP map.
(a) Circular layout of the predicted connections between drugs and cell types (all connections statistically significant P < 0.05 with FDR < 5%, see methods). Line widths correspond to the number of interactions. The diagram has been organized by sorting the cell types counter-clockwise (drugs clockwise) in order of decreasing number of connections. Single letter codes for each drug follows the anatomical therapeutic classification system as described in Fig. 2. (b) Subnetwork showing the drug hubs and islands (square nodes in the center and periphery respectively), and their predicted interactions with immune cell subsets (circles). Each square node represents a single drug (for example, the four center nodes labeled L represent, counter clockwise from top, the drugs daunorubicin, azacitidine, vorinostat, and methotrexate). (c) Drugs identified in the IP map based on their immunemod score and predicted to increase neutrophil and monocyte frequencies in the blood. (d) Validation of drug prediction in immune cells from humans. Drug influence (+ = drug, − = no drug) on measured cell counts from lab values of patients in the Mount Sinai Electronic Medical Records (cell counts and frequencies). Bar heights represent median count (neutrophil) or frequency (monocyte) values and bottom/top of the error bars reflect the inter-quartile ranges (25% to 75% of data respectively). Significance assessed via Wilcoxon rank sum test.