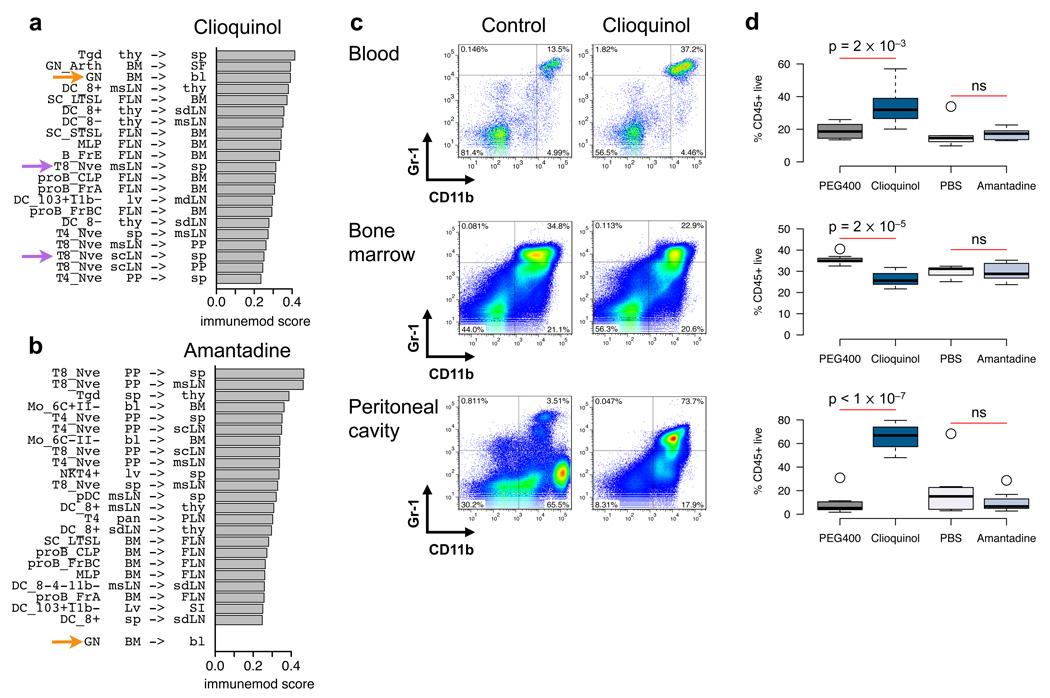
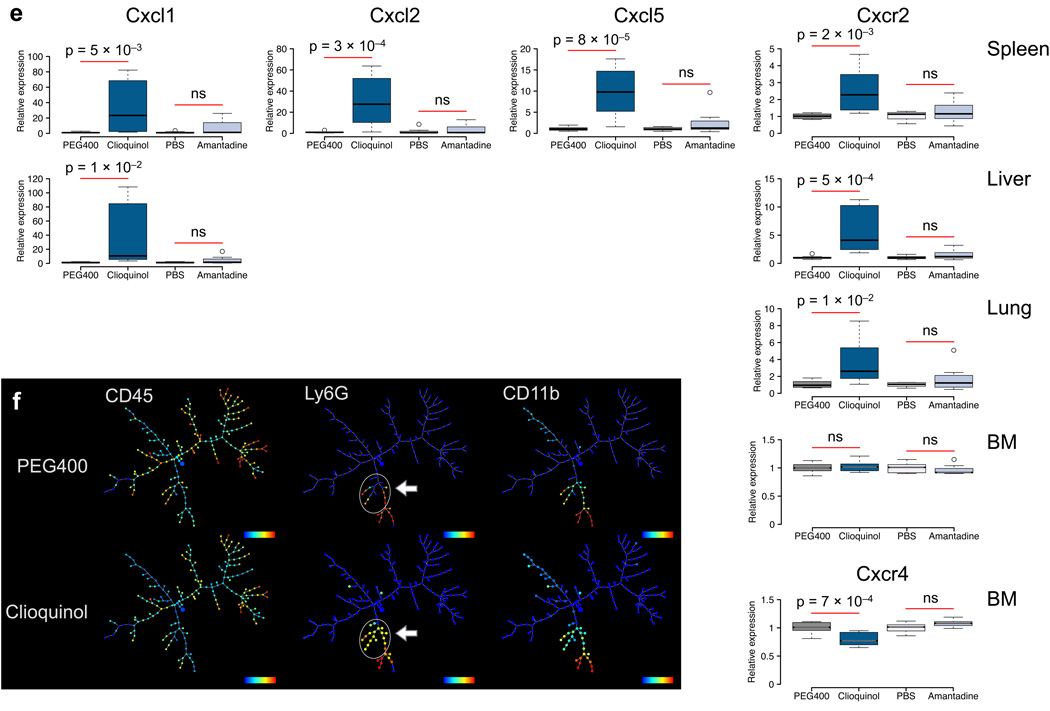
Figure 4. Influencing immune cell migration using the IP map.
Immunemod scores for influencing immune cells between tissues using (a) clioquinol or (b) amantadine. Labels indicate the direction of influence for the drug to a cell type between tissues x -> y, and bar width is the magnitude of the immunemod score. Shown are the significant and robust drug-cell interactions. Orange arrows reflect predicted top interaction with clioquinol and non-interaction with amantadine. Purple arrows point to predicted interactions with additional experimental validation. (c) Representation biaxial flow cytometry plots of Gr-Ly6G+ and CD11b+ cells collected from various tissues of treated and untreated mice. Cells were gated on CD45+DAPI− population. (d) Frequencies of Ly6G+CD11b+ cells neutrophils collected from blood, bone marrow, and the peritoneal cavity (n = 8 per group). Black bars reflect the median expression of all samples in each condition and the bottom/top of the boxes represent the 25th and 75th percentiles respectively. Data are representative of three to four experiments. Significance assessed by ANOVA. (e) Quantitative analysis of neutrophil marker genes expression. The abundance of each transcript in spleen, liver, lung and bone marrow (BM) was calculated relative to the appropriate control (PEG400 for clioquinol or PBS for amantadine). Each experimental condition includes 8 samples. Significance assessed by ANOVA. (f) SPADE trees of CyTOF data collected from cells in spleen. Arrows highlight Gr-Ly6G+CD11b+ population increase after clioquinol treatment. Horizontal color scales represent median intensity for marker indicated (blue = low, red = high).