Figure 3.
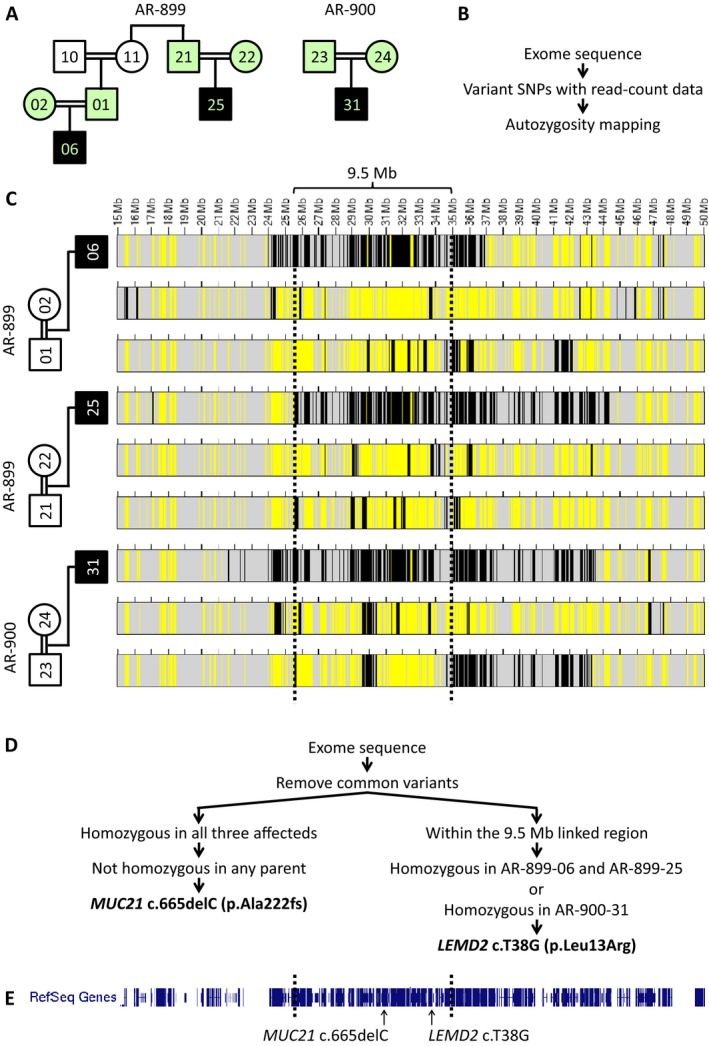
Exome sequencing of three trios enables autozygosity mapping and identification of candidate variants. (A) Exome sequencing was performed on three individuals with juvenile cataracts and their parents (mint shading). (B, C) Regions of homozygosity (black) and heterozygosity (yellow) were determined from exome variant calls using AgileVariantMapper software (Carr et al. 2013). Dotted lines demarcate a 9.5‐Mb autozygous region on chromosome 6 (6p22.2‐p21.31) shared by all three affected individuals and absent from their healthy parents, the only such region in the genome (see Fig. S1). (D) To identify candidate mutations, exome data were first filtered to include only homozygous variants unique to all three affected individuals (consistent with autosomal recessive inheritance). A single variant, MUC21 c.665delC, satisfied these criteria. To identify variants that may have evaded sequencing in one or more samples, particularly as the AR‐899 and AR‐900 samples were captured on different platforms (AR‐899 samples were acquired several years earlier), we performed a parallel filtering scheme to identify homozygous variants within the 9.5 Mb linked region in either the AR‐899 affecteds or AR‐900‐31. This strategy yielded LEMD2 c.T38G, a variant identified only in AR‐900‐31. (E) Both MUC21 and LEMD2 fall within the 9.5 Mb linked region on chromosome 6p.
