Abstract
Small-molecule inhibitors of AKT signaling are being evaluated in patients with various cancer types, but have so far proven therapeutically disappointing for reasons that remain unclear. Here, we treat cancer cells with sub-therapeutic doses of Akti-1/2, an allosteric small molecule AKT inhibitor, in order to experimentally model pharmacologic inhibition of AKT signaling in vitro. We then apply a combined RNA, protein, and metabolite profiling approach to develop an integrated, multi-scale, molecular snapshot of this “AKTlow” cancer cell state. We find that AKT-inhibited cancer cells suppress thousands of mRNA transcripts, and proteins related to the cell cycle, ribosome, and protein translation. Surprisingly, however, these AKT-inhibited cells simultaneously up-regulate a host of other proteins and metabolites post-transcriptionally, reflecting activation of their endo-vesiculo-membrane system, secretion of inflammatory proteins, and elaboration of extracellular microvesicles. Importantly, these microvesicles enable rapidly proliferating cancer cells of various types to better withstand different stress conditions, including serum deprivation, hypoxia, or cytotoxic chemotherapy in vitro and xenografting in vivo. These findings suggest a model whereby cancer cells experiencing a partial inhibition of AKT signaling may actually promote the survival of neighbors through non-cell autonomous communication.
Keywords: cancer, multi-scale profiling, quiescence, microvesicles, stress resistance
INTRODUCTION
Most human cancers activate the AKT kinase signaling pathway either directly through somatic mutation of PTEN, PI3 kinase, or AKT itself, or indirectly through the activation of intersecting oncogenic pathways (1–3). In turn, the AKT kinase activates myriad downstream targets that promote tumor growth, survival, and progression (1). Therefore, most human tumors are thought to depend on AKT signaling to a varying degree for their viability. Based on these observations, AKT-selective small molecule inhibitors have been developed and are currently being evaluated as cancer therapeutics for patients with many different types of malignancy. In pre-clinical xenograft models, however, many AKT inhibitors produce tumor stasis instead of regression (4–6). Moreover, rare patients treated with these inhibitors will occasionally show a significant clinical response to small-molecule AKT inhibition, but most either have partial or minimal responses regardless of PTEN / PI3K / AKT tumor mutational status for reasons that remain unclear (7, 8).
We recently discovered that epithelial cancer cells growing in culture occasionally divide asymmetrically by suppressing AKT signaling in one emerging “AKTlow” daughter cell (9, 10). This unusual type of cell division is triggered by an asymmetric decrease in Type I collagen-β1-integrin-FAK signaling, resulting in activation of the mTORC2 signaling complex, partial phosphorylation of AKT1 kinase, and its activation-induced degradation mediated by the E3-ubiquitin ligase TTC3 and the proteasome (9). Asymmetric signaling thus produces one normally proliferating daughter cell and another AKT1low daughter expressing a MCM2low, H3K9me2low, HES1high marker profile (9, 10). Importantly, suppression of AKT signaling is both necessary and sufficient to produce these slow proliferators (9). AKT1low cancer cells are not apoptotic, autophagic, or senescent, nor do they express cancer stem cell markers or differentiate (10). Rather, they are quiescent but able to eventually resume their cell cycle after a prolonged period of dormancy in vitro (i.e., ~ 7–10 days) (10).
Interestingly, we have also found that human cancer cell lines treated with allosteric small-molecule AKT inhibitors (e.g., Akti-1/2, MK-2206), at a sub-therapeutic dose (i.e., which only partially suppresses AKT signaling by about 80–90%), dramatically increase their fraction of AKTlow, MCM2low, H3K9me2low, HES1high cancer cells (6, 9–11). These quiescent cancer cells rapidly resume their cell cycle with inhibitor washout, consistent with a temporary rather than permanent cell cycle arrest, which is identical to spontaneously arising AKTlow slow proliferators (10). In fact, malignant cells of various types can be made quiescent this way regardless of their PTEN / PI3K / AKT mutation status or general dependency on PI3K / AKT signaling pathway for their growth (9). Based on these observations, we sought to understand this AKT-induced quiescent cancer cell state in further molecular detail using a combined RNA, protein, and metabolite profiling approach to develop an integrated, multi-scale, molecular snapshot of small molecule AKT inhibition.
MATERIALS AND METHODS
Experimental Methods
Cell lines
HCT116 colon, MCF7 breast, MDA-MB-231 breast, A375 melanoma, and PC9 lung were purchased from ATCC, were they were validated. HCT116 AKT1/2−/− was purchased from Horizon Discovery (Cambridge, UK), where it was validated. AG11726 skin fibroblasts were purchased from Coriell Repositories, where they were validated. MCF7, MDA-MB-231 and AG11726 were maintained in DMEM, 10% FCS, 40mM glutamine, 100 U/mL penicillin, and 100μg/mL streptomycin; HCT116 and HCT116 AKT1/2−/− in McCoy’s 5α medium supplemented with 10% FCS, 100U/mL penicillin, and 100μg/mL streptomycin; PC9 in RPMI, 25% glucose, 1% sodium pyruvate, 100U/mL penicillin, and 100μg/mL streptomycin; A375 in DMEM supplemented with high glucose HEPES buffer, 10% FCS, 100U/mL penicillin, and 100μg/mL streptomycin. All the cells were grown at 37°C and 5% CO2.
Induction of AKTlow cancer cells in vitro
Cells were treated for 72h with vehicle (DMSO), Akti-1/2 inhibitor (HCT116: 20μM; MCF7: 2μM; MDA-MB-231: 20μM; A375: 20μM; PC9: 20μM) (Sigma) or MK-2206 (HCT116: 10μM; MCF7: 3μM; MDA-MB-231: 5μM; A375: 10μM; PC9: 3μM) (Selleckchem).
Induction of AKTlow cancer cells followed by xenografting in vivo
HCT116 and MCF7 were treated for 72h with vehicle (DMSO) and Akti-1/2 inhibitor; 500,000 cells were injected subcutaneously into the flanks of 5–6 week old, female, immunocompromised NU/NU mice (Charles River Laboratories), and then growing tumors were measured weekly by caliper.
GRO-sequencing (global run-on)
HCT116 or MCF7 were treated with DMSO and Akti-1/2 for 72h and cells were collected. Isolation of nuclei and nuclear run-on was carried out as described previously (8). Nascent RNAs were on average approximately 100nt long. The immuno-purified RNA was resuspended in 8.5μl water and 5′- or 3′-adapters ligated using Tru-Seq Small RNA Kit, Illumina. RNAs were reverse transcribed and amplified. The NRO-cDNA libraries were then run on a non-denaturing 1XTBE, 8% acrylamide gel, and cDNAs greater than 90 nucleotides were excised from the gel and eluted, precipitated and sequenced on the Illumina HiSeq 2000 Sequencing System.
RNA-Sequencing
We created a dUTP strand-specific cDNA library for RNA-Seq. Total RNA was purified for all the above experiments using RNeasy Mini Kit (Qiagen), and RNA integrity was checked using RNA 6000 Nano Kit on Agilent 2100 Bioanalyzer. Akti-1/2 treated cells showed only a mild decrease (e.g., ~10%) in total RNA concentration compared to DMSO treated cells (i.e., MCF7 DMSO - 38.7μg; MCF7 Akti-1/2 - 35.69μg; HCT116 DMSO - 45.08μg; HCT116 Akti-1/2 - 40.3μg). We used 4μg of total RNA for library construction. The purification, fragmentation and first strand synthesis were performed as described in the Illumina TruSeq RNA Library Prep Kit v2. The second strand cDNA synthesis was modified using the dUTP second strand method (12). End repair, 3′ adenylation and adapter ligation steps were done using TruSeq protocol. The libraries were validated using a High Sensitivity DNA Kit on Agilent 2100 Bioanalyzer, and sequenced using 1 lane of 101bp (for batch 1), or 51bp (for batch 2) paired end reads with the Illumina HiSeq 2000 Sequencing System.
Quantitative Proteomics
We used tandem mass tag reagents (TMT; Thermo Scientific) and a synchronous precursor selection-based MS3 method on an Orbitrap Fusion mass spectrometer (Thermo Scientific) as described previously (13).
Antibody array profiling
MCF7 and HCT116 were treated with DMSO and Akti-1/2 for 6 days and culture supernatant were screened for secreted proteins using the RayBiotech L-Series Human Antibody Array 493 and 507 biotin label-based kits (RayBiotech).
Immunofluorescence staining
Cells were grown directly on collagen IV-coated coverslips. Cells were fixed in 3.7% formalin, permeabilized using 0.1% Triton X-100, and treated with 0.1% SDS. They were blocked in 1% BSA and then incubated with primary antibody (MCM2, Cell Signaling, H3K9me2, Abcam, CD63 (H-193), Santa Cruz, FDFT1, Abcam), followed by the respective secondary antibody, Alexa Fluor conjugates (Invitrogen). Cells were mounted using hard-set mounting media containing DAPI (Vector Laboratories). Cells were stained with Filipin (Sigma) for cholesterol and Alexa 594-conjugated CTXB (Invitrogen) for lipid rafts. Immunofluorescence imaging was performed on a Nikon Eclipse Ti A1R-A1 confocal microscope.
Western blots
We used standard protocols for SDS-PAGE electrophoresis and used the following primary antibody: CD63 (H-193) and CD81 (H-121), Santa Cruz; Calnexin, GM130 and TNFSF10, Abcam. Microvesicle fractions used for the Western Blots were isolated from equivalent number of cells (1×106).
Microvesicle extraction from cell media
Extracellular microvesicles (30–120nm) were isolated from the media of cells treated with DMSO, Akti-1/2 or MK-2206 for 72h as per instructions using the Total Exosome Isolation Reagent (Life Technologies). Cells were cultured in exosome-free media (complete media containing Exosome-depleted FCS). Microvesicle pellets were re-suspended in 100μl PBS.
RNA isolation from microvesicles
Exosomal RNA was isolated as per manufacturer’s instructions using the Total Exosome RNA and Protein Isolation Kit (Life Technologies). Recovered RNA was characterized using Agilent’s RNA 6000 Pico Kit on an Agilent 2100 Bioanalyzer.
Small RNA sequencing
The libraries of cellular and microvesicular small RNA were made using Illumina’s TruSeq Small RNA Kit. The 3′ and 5′ adaptors were ligated, and an RT reaction was used to create single stranded cDNA, which was subsequently PCR amplified using a common primer and one index sequence before size selection on 6% native polyacrylamide gel. Fragment range of 105–150bp, corresponding to the small RNA population, were excised, eluted, precipitated, and resuspended in 20μl of nuclease-free water. The size, quality and quantity of the DNA in each final small RNA library were verified using the High Sensitivity DNA Kit (Agilent).
In vitro cell survival assays with microvesicles
Microvesicles were incubated with recipient cells for 1h at 37°C. Pre-conditioned cells were analyzed for growth for 120h, different stress conditions including growth in 1% fetal calf serum supplemented media, low oxygen (4%), and paclitaxel for 72h. The total number of cells was counted in triplicates. The standard MTS assays were also done for the growth curves. For colony formation assays, cells were treated with microvesicles for 1h and seeded at a density of 400 cells per well in six-well plates, allowed to attach overnight. Cells were then incubated under different stress conditions for an additional 6 days. Colonies were fixed and stained with Coomassie blue and counted in triplicates. In the long-term experiment, these cells were passaged for two additional weeks and then challenged with stress conditions. In addition, microvesicles from the parent cell lines were also incubated with cell lines of different cancer models and skin fibroblasts before exposing them to different stress conditions.
In vivo xenograft tumors with microvesicles
1×105 cells were incubated with microvesicles derived from equivalent number of cells (1×106) for 1h at 37°C. Cells were then injected subcutaneously into the flanks of 5–6 week old, female immunocompromised NU/NU mice (Charles River Laboratories), and the growing tumors were measured weekly by caliper.
Tumor immunohistochemistry
For immunohistochemistry, 5μm sections of formalin-fixed paraffin-embedded (FFPE) tissues were dewaxed with xylene and rehydrated. Antigen retrieval was achieved by microwaving in unmasking solution (Vector Laboratories). After washing, sections were blocked in 5% FCS and immunoperoxidase analysis was performed on tumors for MKI67 (Abcam). The slides were counterstained lightly with hematoxylin for viewing negatively stained cells. H&E slides were used to assess the morphological integrity and geographical variation in morphology of the tissue samples. The slides were analyzed under the LEICA DC500 microscope. Three 40× high-power fields were analyzed for each slide.
Computational Methods
GRO-Seq analysis
Single-end reads were 50bp long. Clipping of contaminating adapter sequences was done with Cutadapt v1.2.1 (14). Reads with poor overall quality were further removed with fastq_quality_filter v0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/). Reads were aligned to the hg19 human genome with Bowtie v0.12.9 (15), allowing no mismatches and discarding multiple mapping reads. An average of ~30M of uniquely aligned reads per sample was obtained. Stranded read counts were obtained using coverageBed v2.17.0 (16). In order to specifically quantify the amount of actively elongated polymerase, the expression level of each gene was quantified based on the number of reads mapping to the region starting at +500 bp downstream the transcription start site (TSS) up to the transcription end site (TES). Genes less than 1 kb long were considered too short to reliably estimate their expression levels and thus were removed from further analyses. Reads per kilobase per million reads (RPKMs) were estimated based on the number reads sequenced per sample, the number of reads mapping to each gene, and the gene mappable length. Each cell line and condition was done in duplicate. Log2-RPKM correlation levels between replicates were between 0.71 and 0.85.
RNA-Seq analysis
Paired-end reads were either 101bp long (1st batch of replicates) or 51bp long (2nd batch of replicates). Clipping of contaminating adapter sequences and trimming of low-quality read ends was done with Trimmomatic v0.25 (6). Average fragment length and its standard deviation were empirically determined for each sample with Bowtie v2.1.0. Hg19/GRCh37 (Feb. 2009) transcriptome was obtained from table “knowGene” in the UCSC Table Browser website (http://genome.ucsc.edu/cgi-bin/hgTables). Tophat v2.0.8b (5) was used to align the reads to hg19 version of the human genome and transcriptome. Multiple mapping reads were excluded from subsequent analyses. Human UCSC hg19 genome annotation was downloaded from Illumina’s FTP repository (ftp://igenome:G3nom3s4u@ussd-ftp.illumina.com/Homo_sapiens/UCSC/hg19/Homo_sapiens_UCSC_hg19.tar.gz) on Feb 21, 2013, corresponding to the UCSC freeze of March 9, 2012. At least 20M uniquely aligned reads were obtained for each sample. Fragments per kilobase per million reads (FPKMs) estimations for each annotated genomic feature were obtained with Cufflinks v2.1.1 (17). Ribosomal, mitochondrial and transfer RNAs were masked in the analysis. Each cell line and condition was done in duplicate. Log2-FPKM correlation levels between replicates were above 0.9.
Quantitative proteomic analysis
Data analysis was done on an in-house developed software suite. MS2 spectra were assigned using the SEQUEST algorithm to search against the human UniProt protein sequence database using a target-decoy database search approach allowing to filter peptide and protein assignments to false-discovery rate of less than 1% (2, 3). MS3 spectra were used for peptide quantification only if the summed signal-to-noise ratios of all 8 TMT ions was greater than 310 and the proportion of non-target ions in the isolation m/z window applied for isolating the target ion was less than 25%. For protein quantification the TMT ion intensities for each TMT channel from each peptide assigned to a protein were summed up and the protein TMT intensities were normalized based on the median TMT intensities of the TMT channel intensities from the pooled standard peptide mixtures (TMT-126 and TMT-131). Each cell line and condition was done in triplicate. Correlation of protein abundance in log2 space between replicates was around 0.8.
Metabolomics analysis
Metabolites were identified by automated comparison in the experimental samples to a reference library of chemical standard entries developed at Metabolon, Inc. Each sample was profiled six times. Most correlation coefficients between replicate pairs fell within the 0.8–0.9 range.
Secreted protein analysis
RayBiotech provided background-subtracted, positive-control normalized intensities for both L-507 and L-493 antibody arrays. For each one of the sub-arrays, all samples were mean-centered to a log2 value of 4.5. Data from both sub-arrays were combined after normalization.
Cellular and microvesicle small RNA-Seq analysis
Single-end reads were 50bp long. Reads with poor overall quality were further removed from downstream analyses with fastq_quality_filter tool. Clipping of contaminating adapter sequences was done with Cutadapt v1.2.1 (14). Reads were aligned to the hg19 human genome (GRCh37.p13) with Bowtie v1.0 (15), allowing no mismatches and discarding multiple mapping reads. More than 20M uniquely aligned reads were obtained for the cellular samples and between 1–3M for the microvesicle libraries. For every annotated feature in the GENCODE v19 database (http://www.gencodegenes.org/releases/19.html), read counts were obtained using the HTSeq Python package (7). Reads normalized per million sequenced reads (RPMs) were subsequently estimated based on the number of million reads sequenced per sample and the number of reads mapping to each gene in that sample. Each cell line and condition was done in duplicate for the cellular samples (biological replicates), while library preparations were sequenced in duplicate for the microvesicle samples (technical replicates). Log2-RPM correlation range between replicates was 0.85–0.99 for the cells and 0.96 for the microvesicles.
Enrichment analyses
All enrichment analyses were computed with GSEA v2.0.14 (18). Paired T-scores comparing AKTi vs DMSO treated samples were used to pre-rank genes. When there were multiple possible pairing combinations, T-scores were computed for all of them and the median T-score was selected. Gene sets with a FWER < 5% were selected as significant. Only canonical gene sets (i.e., KEGG, REACTOME, BIOCARTA, PID, GO) were included in the analyses.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (19) and are accessible through GEO Series accession number GSE71901 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE71901).
RESULTS
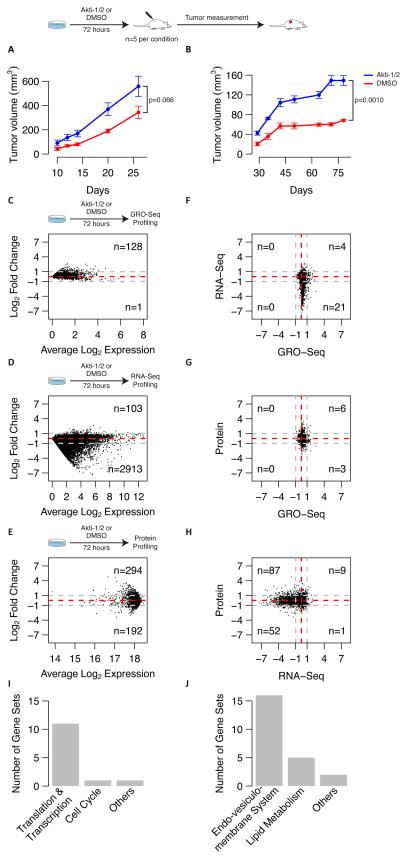
We first treated HCT116 (colon) or MCF7 (breast) cells with a low, non-lethal, cytostatic dose of the allosteric inhibitor Akti-1/2 for three days in vitro, which partially suppresses AKT kinase activity, to induce the AKTlow quiescent cell state rather than killing these cells (i.e., HCT116 = 20μM; MCF7 = 2μM) (10). We then injected 5×105 pre-treated cancer cells subcutaneously in nude mice and assessed their ability to form tumors compared to vehicle-treated control cells (i.e., DMSO, n=5 mice per condition). We previously had found that Akti-1/2 inhibitor treatment increases the fraction of AKTlow slow proliferators within these cancer cell populations from a baseline of 1% up to 60% within 3 days of treatment (10). Surprisingly, we found that transiently increasing the fraction of AKTlow cells, by suppressing AKT signaling in this way, resulted in a substantially improved engraftment of these poorly tumorigenic cell lines compared to DMSO-treated control cells (Fig. 1A–B). This result was counterintuitive since both HCT116 and MCF7 have activating mutations in PIK3CA, rendering them constitutively dependent on AKT signaling for their proliferation, growth, and survival (4).
Figure 1.
A–B: Tumorigenicity of Akti-1/2 treated cells. Plots depict tumor growth curves after injection of 500,000 HCT116 (A) or MCF7 (B) cells in two sets of mice. Blue curves correspond to mice that were injected with Akti-1/2 treated cells, while red curves correspond to mice that were injected with DMSO treated cells. Five mice were initially used for each cell line and condition. Each data point represents the average of the replicates and error bars show the standard error of the mean (SEM). P-values correspond to the t-test statistical differences in tumor volume on the last day of follow-up.
C–E: Multi-scale genomics profiling of AKTlow cells. Average M-A plots for GRO-Seq (A), RNA-Seq (B) and protein (C) datasets for both HCT116 and MCF7 cell lines. X-axis shows the average log2 expression, and y-axis shows the average log2 fold-change between both conditions (Akti-1/2 - DMSO). Positive log2 fold-changes correspond to overexpression in Akti-1/2 treated cells compared to DMSO. Only genes or proteins displaying consistent changes after Akti-1/2 treatment (i.e. log2 fold change either positive or negative in both cell lines) are shown. Numbers on the right side of the plots depict the number of genes or proteins with an average fold change larger than 2-fold (i.e. 1 in log2 space) in absolute terms (grey dashed lines).
F–H: Correlation between changes in each data type. Scatterplots of log2 fold changes for genes or proteins in common between the different data types: GRO-Seq vs RNA-Seq (D), GRO-Seq vs protein (E), RNA-Seq vs protein (F). For each plot, only genes or proteins that show consistent change in both cell lines for both data types are selected. Numbers in the corners correspond to the number of genes or proteins that show a log2 fold change outside the 2-fold change region (grey dashed lines).
I–J: Gene-set enrichment analysis of proteomics results. Barplots depict the number of significantly down-regulated (I) of up-regulated (J) gene sets in AKTlow cells that fall in each functional category (FWER < 5%). Only canonical gene sets (i.e., KEGG, REACTOME, BIOCARTA, PID, GO) were included in these analyses. Complete lists of gene sets queried can be found in Supplementary dataset S2.
To determine the basis of this paradox, we next applied a battery of RNA, protein, and metabolite profiling technologies to Akti-1/2-treated cells in order to define the AKTlow cell state in further molecular detail. We performed: 1) genome-wide GRO-sequencing (global run-on) to examine active transcription across the genome; 2) RNA-sequencing to measure genome-wide steady-state mRNA levels; 3) multiplexed, quantitative mass spectrometry-based proteomics to assess levels of approximately 10,000 proteins at steady state; and 4) mass spectrometry-based metabolite profiling to assess levels of approximately 375 metabolites at steady state. Integration of these datasets allowed us to define a multi-scale molecular snapshot of AKT-inhibited cell quiescence (Fig. 1C–H). Details on experimental procedures, data quality, bioinformatics, and computational analyses for these different data types can be found in the methods section. Complete results for each dataset can be found in Supplementary dataset S1.
We first compared transcriptional profiles from AKTi-treated and DMSO-treated control cells. We focused on transcripts or proteins with greater than an average 2-fold change after AKTi treatment in both HCT116 and MCF7 cells. We found that AKTi-treated cells displayed only a subtle increase in the expression of a few transcripts compared to control cells at the GRO-Seq level (n = 128) (Fig. 1C and Supplementary dataset S1A). Furthermore, gene-set enrichment analysis (i.e., GSEA) applied to this GRO-Seq profile did not reveal significant enrichment in any gene sets out of the 2000+ that we tested (FWER < 5%) (18). In addition, computational analysis of GRO-Seq profiles comparing pausing indexes for all genes across conditions also failed to reveal global changes transcriptional activity in AKTi-treated versus control cells (Supplementary Fig. S1). These results suggested that AKTi-induced quiescence was likely not associated with programmatic changes in RNA PolII-associated transcriptional activity.
Consistent with these findings, RNA-Seq profiling further confirmed that AKTlow cancer cells did not up-regulate many transcripts at steady-state (Fig. 1D and Supplementary dataset S1B). In contrast, however, slow proliferators suppressed thousands of mRNAs at steady state compared to rapidly proliferating cells (i.e., 2913 genes < -2-fold across both HCT116 and MCF7 cells, ~16% of total number of profiled transcripts) (Fig. 1D). However, most of the transcripts beyond this threshold (i.e., 85% - 2483/2913) showed mild expression levels (i.e., average log2 expression < 4), while many of the higher expressed genes that account for most of the sequenced reads remain stable after Akti-1/2 treatment. GSEA applied to this down-regulated RNA-Seq signature, however, only revealed statistically significant enrichment in a single gene set related to XBP1-mediated protein folding (FWER < 5%) (Supplementary dataset S2A). Moreover, this mRNA suppression did not relate to decreases in the active transcription of these genes as determined by GRO-seq analysis (Fig. 1F). Overall, these results were consistent with AKT inhibition producing a global, post-transcriptional, and largely random degradation of many mRNA transcripts.
Against this transcriptional backdrop, we identified 192 proteins that were also down-regulated after AKTi treatment in both HCT116 and MCF7 cells (i.e., < -2-fold) (Fig. 1E). For some of these proteins, corresponding mRNA transcripts were also suppressed, while for others we found that protein and mRNA levels correlated poorly, suggesting a mixed transcriptional and post-transcriptional effect (Fig. 1H). Moreover, GSEA applied to this down-regulated protein signature was associated with statistically significant enrichments in 13 gene sets related to cell cycle transit, ribosomal activity, and translational regulation (FWER < 5%) (Fig. 1I and Supplementary datasets S2B-C). These findings were consistent with known effects that inhibition of AKT signaling might be predicted to have on cell cycle, ribosome function, and RNA stability (1).
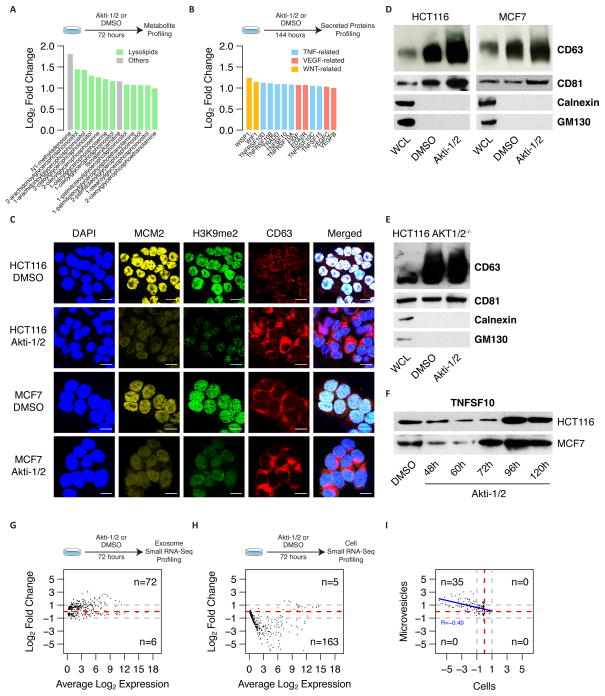
We also identified 294 proteins that were up-regulated after AKTi treatment in both HCT116 and MCF7 cells (> 2-fold) (Fig. 1E). The expression of these proteins correlated poorly with changes in either active transcription or steady state mRNA levels, however, suggesting that these changes more likely related to post-translational protein stabilization (Fig. 1G–H). Furthermore, GSEA of this up-regulated protein signature revealed a significant enrichment in 23 gene sets related to cholesterol biosynthesis, lipid metabolism, the endoplasmic reticulum, vesiculo-membrane transport, trafficking, secretion, along with membrane and extracellular matrix proteins (FWER < 5%) (Fig. 1J and Supplementary datasets S2D-E). Metabolite profiling further suggested the up-regulation of a select set of 13 out of 379 metabolites analyzed, but did not reveal major changes in metabolites related to cellular energetics in AKTi-treated versus control cells (i.e., > an average 2-fold change for both HCT116 and MCF7 cells) (Fig. 2A). Eleven of these thirteen metabolites were lysolipid derivatives that are major components of cell membranes. Furthermore, cholesterol and cholesterol-like molecules (e.g., lathosterol) also showed milder increases in quiescent cells (Supplementary dataset S1D). Consistent with this finding, we also noted the corresponding up-regulation of FDFT1 protein (which is a rate-limiting enzyme in the cholesterol biosynthetic pathway) with proteomic profiling (Supplementary dataset S1C).
Figure 2.
A: Metabolic profiling of AKTlow cells. Barplot depicts the log2 fold-changes (Akti-1/2 - DMSO) of 13 up-regulated metabolites with an average fold change > 2 (log2 > 1). Green bars correspond to lysolipid metabolic derivatives.
B: Secretory profiling of AKTlow cells. Barplot depicts the log2 fold-changes (Akti-1/2 - DMSO) of 13 up-regulated secreted proteins with an average fold change > 2 (log2 > 1). Blue, red, and orange bars correspond to proteins related to the TNF, VEGF and WNT families, respectively.
C: Immunofluorescence images for both HCT116 and MCF7 cells, treated either with DMSO or Akti-1/2. Cells were stained for DAPI, MCM2, H3K9me2 and CD63. Merged images represent respective stains merged with underlying DAPI stain. Size bar = 10μm.
D–E: Western blots of purified microvesicle fractions probed for CD63 and CD68 (i.e., exosome markers), Calnexin (i.e. ER vesicles marker), and GM130 (i.e., Golgi vesicles marker) in HCT116 and MCF7 (D), and HCT116 AKT1/2−/− knockout (E) cell lines after treatment either with DMSO or Akti-1/2 for 72h. A whole cell lysate (WCL) has been added as a control. For each cell line and condition microvesicle fractions were isolated from equivalent number of cells (1×106).
F: Western blots of purified microvesicle fractions probed for TNFSF10 in HCT116 (left panel) and MCF7 (right panel) after treatment either with DMSO or Akti-1/2. Microvesicle fractions used for the Western Blots were isolated from equivalent number of cells (1×106).
G–H: Microvesicle (G) and cell (H) small RNA-Seq profiling. Average M-A plot of both HCT116 and MCF7 cell lines. X-axis shows the average log2 small RNA abundance, and y-axis shows the average log2 fold-change between both conditions (Akti-1/2 - DMSO). Positive log2 fold-changes correspond to a small RNA abundance increase in Akti-1/2 treated cells compared to DMSO. Only small RNAs displaying consistent changes after Akti-1/2 treatment (i.e., log2 fold change either positive or negative in both cell lines) are shown. Numbers on the right side of the plots depict the number of small RNAs with an average fold change larger than 2-fold (i.e. 1 in log2 space) in absolute terms (grey dashed lines).
I: Correlation between changes in microvesicle and cell small RNA abundance. X-axis corresponds to log2 fold change in cellular small RNA expression (Akti-1/2 - DMSO), while y-axis corresponds to log2 fold change in microvesicle small RNA expression (Akti-1/2 - DMSO). Positive log2 fold-changes correspond to a small RNA abundance increase in Akti-1/2 treated cells compared to DMSO-treated cells. Numbers in the corners correspond to the number of small RNAs that show a log2 fold change outside the 2-fold change region (grey dashed lines).
In addition, we used a highly validated antibody array platform to measure the expression of approximately one thousand different cytokines, chemokines, growth factors, and receptors in conditioned media from Akti-1/2-treated HCT116 and MCF7 cells. These experiments suggested the up-regulation of a small set of 13 secreted proteins (Fig. 2B and Supplementary dataset S1E). This set included multiple TNF, VEGF, and WNT family members known to powerfully modulate a spectrum of cell types, including epithelial, mesenchymal, vascular, and immune cells (e.g., TNFSF10) (20). Overall, these results were consistent with the notion that AKTi-induced slow proliferators might broadly activate their endo-vesiculo-membrane system, membrane formation, membrane remodeling, and secretion of bioactive factors. Initial validation experiments using immunofluorescence confocal microscopy confirmed increase in the expression of CD63 (i.e., a strongly up-regulated membrane protein), FDFT1, cell membrane cholesterol, and membrane lipid rafts after Akti-1/2 treatment in HCT116 and MCF7 cells (Fig. 2C and Supplementary Fig. S2).
CD63 is not only expressed on cell membranes but also marks exosomes, which are extracellular microvesicles that are secreted by both cancer and normal cells (21). These microvesicles are known to mediate cell-cell communication within cancer microenvironments through complex mechanisms that have yet to be fully elucidated (22). We therefore asked whether AKTlow cancer cells also increase their secretion of extracellular microvesicles. We used differential solubility to biochemically isolate secreted microvesicles ranging in size from 30–120nm (which includes the CD63/CD81+ exosome fraction) in conditioned media from Akti-1/2 treated HCT116 and MCF7 cells compared to control. These experiments, which compared whole cell lysates to microvesicles isolated from equivalent numbers of either treated or untreated cells, confirmed the increased secretion of CD63/CD81-expressing microvesicles by Akti-1/2-treated cells in HCT116 and MCF7 cell lines. Further immunoblotting for Calnexin (i.e., an ER-vesicle marker) and GM130 (i.e., a Golgi vesicle marker) excluded other potential vesicle contaminants in enriched microvesicle fractions (Fig. 2D). We also isolated microvesicles from the HCT116-AKT1/2−/− cell line, which has adeno-associated virus (AAV)-mediated disruption of the AKT1 and AKT2 gene loci (23). AKT1/2−/− cells do not express either AKT1 or AKT2, nor do they express AKT3, and thus survive and proliferate albeit poorly in the complete absence of AKT signaling, presumably through compensatory changes that arose during their initial selection. Akti-1/2 treatment of these HCT116-AKT1/2−/− cells did not produce an increase in CD63/CD81+ microvesicle secretion as observed with wild-type HCT116 cells (Fig. 2E). Overall, these results supported the idea that AKT signaling negatively regulates microvesicle secretion by cancer cells. In addition, these microvesicles displayed a time-dependent increase in the expression of TNFSF10 (Fig. 2F), which we had identified as up-regulated in conditioned media of Akti-1/2-treated cells using antibody arrays. RNA-sequencing also revealed an increase in the expression of various small RNAs in microvesicles from Akti-1/2-treated compared to control cells (> an average 2-fold change for both HCT116 and MCF7) (Fig. 2G and Supplementary dataset S1F). These same microRNAs were suppressed within AKTi-treated cells themselves, however, suggesting their active export in microvesicles. (R=−0.49) (Fig. 2H–I and Supplementary dataset S1G).
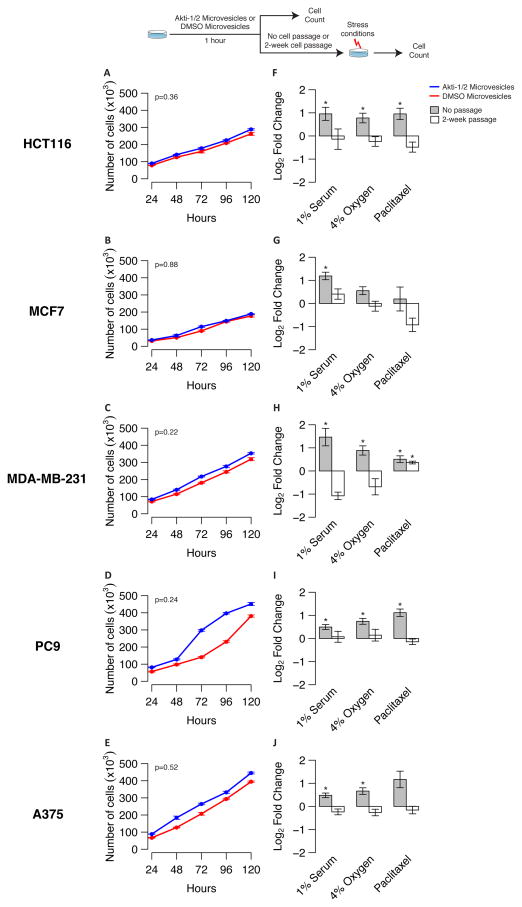
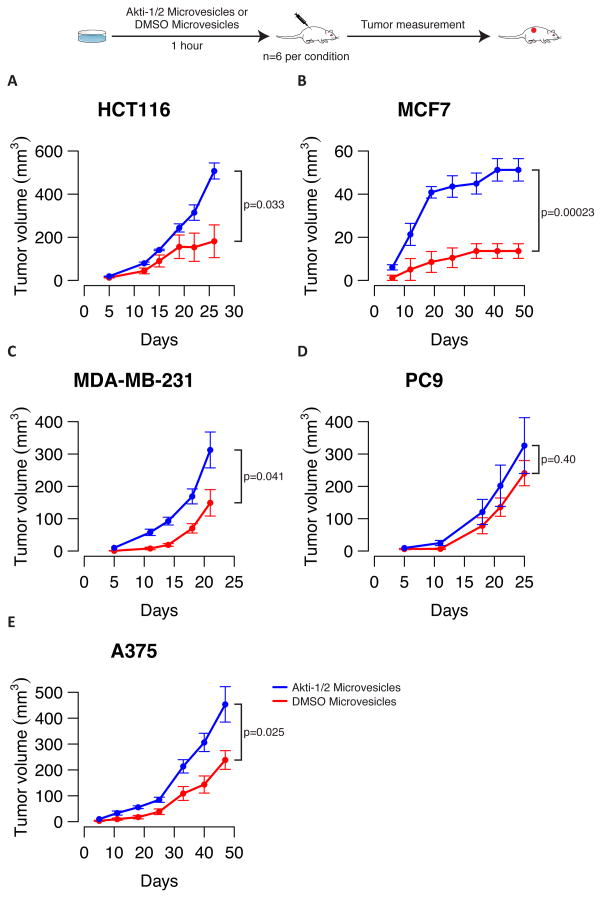
Next, we asked whether microvesicles isolated from AKT-inhibited cancer cells have functional effects either in vitro or in vivo. We used Akti-1/2 inhibition to produce AKTi-induced microvesicles from five different human cancer cell lines of different molecular types (i.e., HCT116, MCF7, A375 (melanoma) (20 μM), MDA-MB-231 (breast) (20 μM), and PC9 (lung) (20 μM)). We then admixed either Akti-1/2-induced or control microvesicles with untreated, isogenic cancer cells for one hour, and examined the behavior of these pre-treated cells in a variety of functional assays in vitro. We found that pre-conditioning with AKTi-induced microvesicles did not increase the growth of target cells in culture over time, as assessed by either direct cell count (Fig. 3A–E) or vitality (i.e. MTS) assay (Supplementary Fig. S3A–E). Based on both cell and colony count assays, pre-treatment with microvesicles derived from AKTi-treated cells mostly increased the resistance of proliferating cancer cells to various stress conditions, however, including in vitro serum deprivation (1%), hypoxia (4%), and paclitaxel chemotherapy (0.05μM, HCT116; 0.5μM, MCF7; 0.001μM, MDA-MB-231; 10μM, PC9 and 0.05μM, A375; and 2.5μM, AG11726) (Fig. 3F–J, Supplementary Fig. S3F–J). Importantly, we also obtained similar results when pre-treating cancer cells with MK-2206, a second allosteric AKT1/2 small molecule inhibitor, further confirming that these effect likely related to small molecule inhibition of AKT signaling (Supplementary Fig. S4). In contrast, cancer cells pre-treated with Akti-1/2 microvesicles, but passaged for two weeks before challenge, did not display an increased resistance to stress, suggesting a transient rather than prolonged effects in vitro (Fig. 3F–J). Additionally, microvesicles from one cell type (e.g., HCT116) could not pre-condition virgin cancer cells of other types (i.e., MCF7, MDA-MB-231, PC9, A375) or normal human fibroblasts (i.e., AG11726) to withstand these stress conditions in vitro (Supplementary Fig. S5). Finally, we exposed cancer cells for 1 hour to microvesicles derived from 1×106 Akti-1/2 or DMSO treated cells. In order to assess xenograft efficiency, we subcutaneously injected 100,000 pre-treated cells in nude mice (n=6, initially per condition). Remarkably, pre-treatment with AKTi-induced microvesicles resulted in the increased engraftment of isogenic cancer cells in most cell lines tested relative to control, which was only associated with small, inconsistent differences in MKI67 expression within resulting experimental and control tumors (Fig. 4 and Supplementary Fig. S6). Similarly, DMSO or Akti-1/2-treated cancer cells also produced tumors displaying inconsistent differences in MKI67 expression (Supplementary Fig. S7). These functional experiments supported a model whereby microvesicles that are secreted by AKT-inhibited slow proliferators can promote the context-specific, non-cell autonomous survival of rapidly proliferating cancer cells exposed to a variety of experimental stresses, including xenotransplantation both in vitro and in vivo.
Figure 3.
A–E: Effect of microvesicles on growth rate. Different tumor cell lines (e.g., HCT116 (A), MCF7 (B), MDA-MB-231 (C), PC9 (D), and A375 (E)) were exposed to microvesicles derived from either Akti-1/2 or DMSO treated cells of the same type for 1 hour. Experiments were done in triplicate for each cell line and treatment. Error bars show the standard error of the mean (SEM). P-values shown at the top-left corner on each panel correspond to the model comparing the two slopes of the linear models fitted for each condition (i.e., DMSO or Akti-1/2 microvesicles). Non-significant p-values in all five cell lines support the finding that there is no evidence of any significant difference in cell growth based on the differential microvesicle treatment.
F–J: Akti-1/2 microvesicles bioactivity in vitro. Barplots depict the log2 fold change (Akti1/2 - DMSO) in the total number of cells exposed to microvesicles derived from either Akti-1/2 or DMSO treated cells of the same type for 1 hour. Cells were then placed under three different stress conditions (e.g. 1% serum, 4% oxygen, standard chemotherapy agent) for 72h (grey bar). After pre-conditioning, cells were also passaged for two weeks before placing under three different stress conditions (e.g. 1% serum, 4% oxygen, standard chemotherapy agent) for 72h (open bar). Experiments were done in triplicates for each 5 different cells line (e.g., HCT116 (F), MCF7 (G), MDA-MB-231 (H), PC9 (I), and A375 (J)), treatment, and stress condition. Error bars show the standard error of the mean (SEM). Asterisks on top of bars designate statistically significant increases (i.e., p < 0.05, one-sided t-test) in cell count after exposing them to microvesicles derived from Akti-1/2 treated cells, compared to microvesicles derived from DMSO treated cells.
Figure 4.
A–E: Akti-1/2 microvesicles bioactivity in vivo. Plots depict tumor growth curves after injection of tumorigenic cells exposed during 1 hour to microvesicles derived from equivalent (i.e., 1×106) numbers of either Akti-1/2 or DMSO treated cancer cells (e.g., HCT116 (A), MCF7 (B), MDA-MB-231 (C), PC9 (D), and A375 (E)). 500,000 cells were injected into each mice. Blue curves correspond to mice that were injected with cells admixed with microvesicles derived from Akti-1/2 treated cells, while red curves correspond to mice that were injected with cells admixed with microvesicles derived from DMSO treated cells. Six mice were initially used for each cell line and condition. Each data point is an average of all replicates and error bars show the standard error of the mean (SEM). P-values correspond to the t-test statistical differences in tumor volume on the last day of follow-up.
DISCUSSION
Small molecule drugs that are designed to target specific aspects of cell behavior may produce unanticipated biological effects that are interesting but might ultimately compromise their therapeutic utility. Strategies to systematically understand these effects may therefore prove valuable for both biological research and pre-clinical drug development. We have piloted a multi-scale profiling approach to functionally assess inhibition of AKT signaling in human cancer cell lines. In these experiments, we use a single, sub-therapeutic drug dose of Akti-1/2, which is a well-studied, prototypic, small molecule, allosteric AKT inhibitor to partially inhibit AKT signaling, and a time of exposure carefully chosen in each individual cell line to induce a reversibly quiescent cell state rather cell death (i.e., 3 days) (10). We specifically focus on allosteric AKT inhibitors in our studies, since we previously found that catalytic AKT inhibitors do not induce the same quiescent cell phenotype, suggesting a class-specific inhibitor effect, likely related to the ability of allosteric but not catalytic inhibitors to induce degradation of AKT protein (24).
We apply combined RNA, protein, and metabolite profiling to this highly validated experimental system in order to develop an integrative molecular view of the AKTlow cell state. This multi-scale profiling strategy reveals a rich and complex landscape of molecular activity in AKT-inhibited cancer cells. Surprisingly, AKT-inhibited cancer cells continue to actively transcribe most genes similar to rapidly proliferating cells, but post-transcriptionally suppress several thousand mRNAs and proteins, consistent with prior observations regarding AKT signaling and its regulation of cell cycle transit, transcript stability, ribosomal activity, and protein translation (1). In addition, AKT-inhibited slow proliferators appear to post-translationally increase their expression of endo-vesiculo-membrane proteins, membrane remodeling, secretion of inflammatory proteins, and elaboration of extracellular microvesicles. We further find that microvesicles from both Akti-1/2 and MK-2206-inhibited cells functionally increase the resistance of a molecularly diverse panel of target cancer cell types to various stress conditions including serum deprivation, hypoxia, and chemotherapy exposure in vitro. Additional experiments will be required to elucidate at a molecular level how precisely microvesicles elaborated in response to AKT-inhibition promote cancer cell survival. Several miRNAs that we identify within Akti-1/2-induced microvesicles have been previously reported to play roles in response to hypoxia (i.e., miR-210 (25, 26)); stress response (i.e., miR-320a (27), miR-574 (28)); and chemotherapy resistance (i.e., miR-92b (29), miR-375 (30, 31), miR-345 (32), miR-197 (33, 34), and miR-140 (35)). It is therefore possible that these or other individual miRNAs promote stress-resistance in trans, and that mild differences in biological effect size that we observe across different cell types might relate to multiple factors including specific microvesicle content or the molecular profiles of target cells. Nevertheless, microvesicles from one cancer cell line do not induce stress resistance when applied to cancer or normal cells of different types in vitro, suggesting additional, complex, and context-specific effects yet to be fully elucidated. Intriguingly, cancer cells of various types, either pre-treated with a sub-therapeutic dose of Akti-1/2, or with microvesicles isolated from Akti-1/2-treated cells, also display increased engraftment upon xenografting into nude mice in vivo. While a transient ability of microvesicle-treated cells to withstand xenotransplantation-associated stress might account for increased experimental tumorigenesis, additional experiments are required to determine the possibility of more sustained effects on tumor growth in vivo.
Cancer cells growing in culture or within tumors continuously encounter both internal (e.g., oncogenic, proteotoxic) and external (e.g., hypoxic) stresses, and use a range of cellular programs to survive this constant pressure (36, 37). We recently discovered that dividing epithelial cancer cells encountering a loss of integrin signaling trigger a conserved mechanism to partially suppress AKT signaling and produce a newborn “AKTlow” daughter cell (9, 10). These “G0-like” daughters remain quiescent for a period of time within the population before eventually resuming their cell cycle (10). Interestingly, we have also identified these AKTlow slow cyclers within actual human breast tumors, where they appear to survive exposure to intensive, prolonged, combination chemotherapy (10). Our current findings suggest that epithelial cancer cells in this AKT-inhibited state may in fact instruct rapidly proliferating neighbors to increase their resistance to stressful challenge. This view might explain why rapid proliferators, which have evolved through years of mutation and clonal selection, continuously produce small fractions of naturally-arising AKTlow slow proliferators via a conserved signaling mechanism (9, 10). Slow proliferators, while only marginally reducing overall population expansion, may not only resist cytotoxic challenge themselves because they are slowly cycling, but might also provide a survival advantage to more rapidly proliferating neighbors. Consistent with this model, recent findings suggest that targeted inhibition of growth factor signaling in cancer cells (i.e., EGFR, HER2, ALK, MET, KRAS) might contribute to the drug resistance of neighboring cells through secreted factors such as IFN-γ (38). Similarly, targeted inhibition BRAF, ALK, or EGFR in cancer cells may induce a complex, reactive secretome that both enhances cancer cell drug resistance and also supports the expansion and dissemination of drug resistant clones in vivo (39). We also note that cells undergoing programmed senescence secrete various inflammatory proteins, most notably IL6 and IL8 (i.e., the senescence-associated secretory phenotype) (40). Unlike senescent or drug tolerant phenotypes, however, AKT-inhibited quiescent cells apparently increase their secretion of different factors including exosomes and WNT-, TNF, and VEGF-related proteins (41). Nevertheless, an emerging body of work suggests that various cell stresses, either naturally arising or iatrogenic, may trigger cell-cell interactions of various types within tumors with potentially important consequences.
Since the non-selective AKT inhibitors that we use inhibit both AKT1 and AKT2 isoforms (i.e., Akti-1/2 and MK-2206), and the HCT116-AKT1/2−/− cells that we use in further validation experiments also lack activity for both these genes, additional experiments will be required to determine whether observed effects relate to inhibition of a single or multiple AKT isoforms. Given that allosteric AKT inhibitors have proven disappointing when used clinically despite strong pre-clinical rationale for their development, however, we speculate that incomplete pharmacologic inhibition of AKT signaling with small molecule AKT inhibitors within tumors may paradoxically increase cancer cell survival through the type of non-cell autonomous communication we observe, rather than inducing cell death as intended (4). Finally, no single type of molecular profiling technology was sufficient to reveal the biological insight provided by our multi-scale approach. For example, RNA-profiling alone would suggest that Akti-1/2 treatment generally results in transcriptional repression, which is not consistent with the fuller picture revealed through a multi-scale profiling approach. Additionally, while proteomic profiling appears to be a richer source of information in this context, suggesting the broad-scale activation of biological pathways related to endo-membrane trafficking, we find that metabolite and secreted protein profiling adds layers of useful information that further sharpen the focus of down-stream biological validation experiments. Additional experiments with related or different drugs, across full dose and time ranges, in many additional cell lines, and using additional profiling technologies (e.g., translational profiling) may therefore prove useful as a general approach that complements ongoing efforts aimed at understanding the molecular action of cancer therapeutics (12, 15).
Supplementary Material
Acknowledgments
FINANCIAL INFORMATION
This work was supported by awards from the National Cancer Institute (R01 CA 185086, C06 CA 05926), Howard Hughes Medical Institute (Physician-Scientist Early Career Award), Susan G. Komen for the Cure, and Prostate Cancer Foundation (to S. Ramaswamy). S. Ramaswamy was supported by a Stand Up To Cancer Innovative Research Grant (SU2CAACR-IRG0411). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. X. Solé was supported by the “Bolsa de Ampliación de Estudios, Instituto de Salud Carlos III, Ministerio de Economía y Competitividad (BA12/00021)” – Spanish fellowship award. C.P. Alves was supported by an award from CNPq “Ciência sem Fronteiras” – Brazil (202620/2012-3). L. Ritsma was supported by a Rubicon Fellowship from the Netherlands Organization for Scientific Research ([NWO] 825.13.016).
We thank Steven Gygi (Harvard Medical School) and Ruslan Sadreyev (Massachusetts General Hospital) for access to computational software and facilities used for processing raw proteomic and genomic data.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods. 2007;4:207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 3.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PloS one. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic acids research. 2012;40:W622–7. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dey-Guha I, Alves CP, Yeh AC, Salony, Sole X, Darp R, et al. A mechanism for asymmetric cell division resulting in proliferative asynchronicity. Molecular cancer research : MCR. 2015;13:223–30. doi: 10.1158/1541-7786.MCR-14-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey-Guha I, Wolfer A, Yeh AC, JGA, Darp R, Leon E, et al. Asymmetric cancer cell division regulated by AKT. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12845–50. doi: 10.1073/pnas.1109632108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nature methods. 2010;7:709–15. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Rechem C, Black JC, Boukhali M, Aryee MJ, Graslund S, Haas W, et al. Lysine demethylase KDM4A associates with translation machinery and regulates protein synthesis. Cancer discovery. 2015;5:255–63. doi: 10.1158/2159-8290.CD-14-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–2. [Google Scholar]
- 15.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–9. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–5. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 21.Kim DK, Lee J, Simpson RJ, Lotvall J, Gho YS. EVpedia: A community web resource for prokaryotic and eukaryotic extracellular vesicles research. Seminars in cell & developmental biology. 2015 doi: 10.1016/j.semcdb.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Webber J, Yeung V, Clayton A. Extracellular vesicles as modulators of the cancer microenvironment. Seminars in cell & developmental biology. 2015 doi: 10.1016/j.semcdb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Ericson K, Gan C, Cheong I, Rago C, Samuels Y, Velculescu VE, et al. Genetic inactivation of AKT1, AKT2, and PDPK1 in human colorectal cancer cells clarifies their roles in tumor growth regulation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2598–603. doi: 10.1073/pnas.0914018107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jo H, Mondal S, Tan D, Nagata E, Takizawa S, Sharma AK, et al. Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10581–6. doi: 10.1073/pnas.1202810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X, Le QT, Giaccia AJ. MiR-210--micromanager of the hypoxia pathway. Trends in molecular medicine. 2010;16:230–7. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JH, Park SG, Song SY, Kim JK, Sung JH. Reactive oxygen species-responsive miR-210 regulates proliferation and migration of adipose-derived stem cells via PTPN2. Cell death & disease. 2013;4:e588. doi: 10.1038/cddis.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, et al. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–66. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa K, Ishikawa A, Shoji Y, Imai T. A genotoxic stress-responsive miRNA, miR-574-3p, delays cell growth by suppressing the enhancer of rudimentary homolog gene in vitro. International journal of molecular sciences. 2014;15:2971–90. doi: 10.3390/ijms15022971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Li L, Guan Y, Liu X, Meng Q, Guo Q. MiR-92b regulates the cell growth, cisplatin chemosensitivity of A549 non small cell lung cancer cell line and target PTEN. Biochemical and biophysical research communications. 2013;440:604–10. doi: 10.1016/j.bbrc.2013.09.111. [DOI] [PubMed] [Google Scholar]
- 30.Shen Y, Wang P, Li Y, Ye F, Wang F, Wan X, et al. miR-375 is upregulated in acquired paclitaxel resistance in cervical cancer. British journal of cancer. 2013;109:92–9. doi: 10.1038/bjc.2013.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Y, Zhou J, Li Y, Ye F, Wan X, Lu W, et al. miR-375 mediated acquired chemo-resistance in cervical cancer by facilitating EMT. PloS one. 2014;9:e109299. doi: 10.1371/journal.pone.0109299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schou JV, Rossi S, Jensen BV, Nielsen DL, Pfeiffer P, Hogdall E, et al. miR-345 in metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PloS one. 2014;9:e99886. doi: 10.1371/journal.pone.0099886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita Y, Yagishita S, Hagiwara K, Yoshioka Y, Kosaka N, Takeshita F, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Molecular therapy : the journal of the American Society of Gene Therapy. 2015;23:717–27. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou D, Wang D, Li R, Tang Y, Yuan L, Long X, et al. MiR-197 induces Taxol resistance in human ovarian cancer cells by regulating NLK. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015 doi: 10.1007/s13277-015-3365-7. [DOI] [PubMed] [Google Scholar]
- 35.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–74. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature reviews Cancer. 2005;5:761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 38.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer cell. 2014;26:207–21. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 39.Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–72. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annual review of pathology. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Analytical chemistry. 2012;84:7469–78. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.