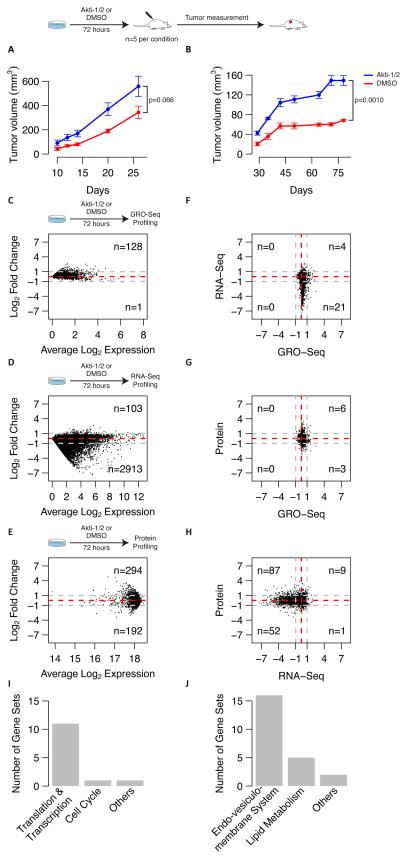
Figure 1.
A–B: Tumorigenicity of Akti-1/2 treated cells. Plots depict tumor growth curves after injection of 500,000 HCT116 (A) or MCF7 (B) cells in two sets of mice. Blue curves correspond to mice that were injected with Akti-1/2 treated cells, while red curves correspond to mice that were injected with DMSO treated cells. Five mice were initially used for each cell line and condition. Each data point represents the average of the replicates and error bars show the standard error of the mean (SEM). P-values correspond to the t-test statistical differences in tumor volume on the last day of follow-up.
C–E: Multi-scale genomics profiling of AKTlow cells. Average M-A plots for GRO-Seq (A), RNA-Seq (B) and protein (C) datasets for both HCT116 and MCF7 cell lines. X-axis shows the average log2 expression, and y-axis shows the average log2 fold-change between both conditions (Akti-1/2 - DMSO). Positive log2 fold-changes correspond to overexpression in Akti-1/2 treated cells compared to DMSO. Only genes or proteins displaying consistent changes after Akti-1/2 treatment (i.e. log2 fold change either positive or negative in both cell lines) are shown. Numbers on the right side of the plots depict the number of genes or proteins with an average fold change larger than 2-fold (i.e. 1 in log2 space) in absolute terms (grey dashed lines).
F–H: Correlation between changes in each data type. Scatterplots of log2 fold changes for genes or proteins in common between the different data types: GRO-Seq vs RNA-Seq (D), GRO-Seq vs protein (E), RNA-Seq vs protein (F). For each plot, only genes or proteins that show consistent change in both cell lines for both data types are selected. Numbers in the corners correspond to the number of genes or proteins that show a log2 fold change outside the 2-fold change region (grey dashed lines).
I–J: Gene-set enrichment analysis of proteomics results. Barplots depict the number of significantly down-regulated (I) of up-regulated (J) gene sets in AKTlow cells that fall in each functional category (FWER < 5%). Only canonical gene sets (i.e., KEGG, REACTOME, BIOCARTA, PID, GO) were included in these analyses. Complete lists of gene sets queried can be found in Supplementary dataset S2.