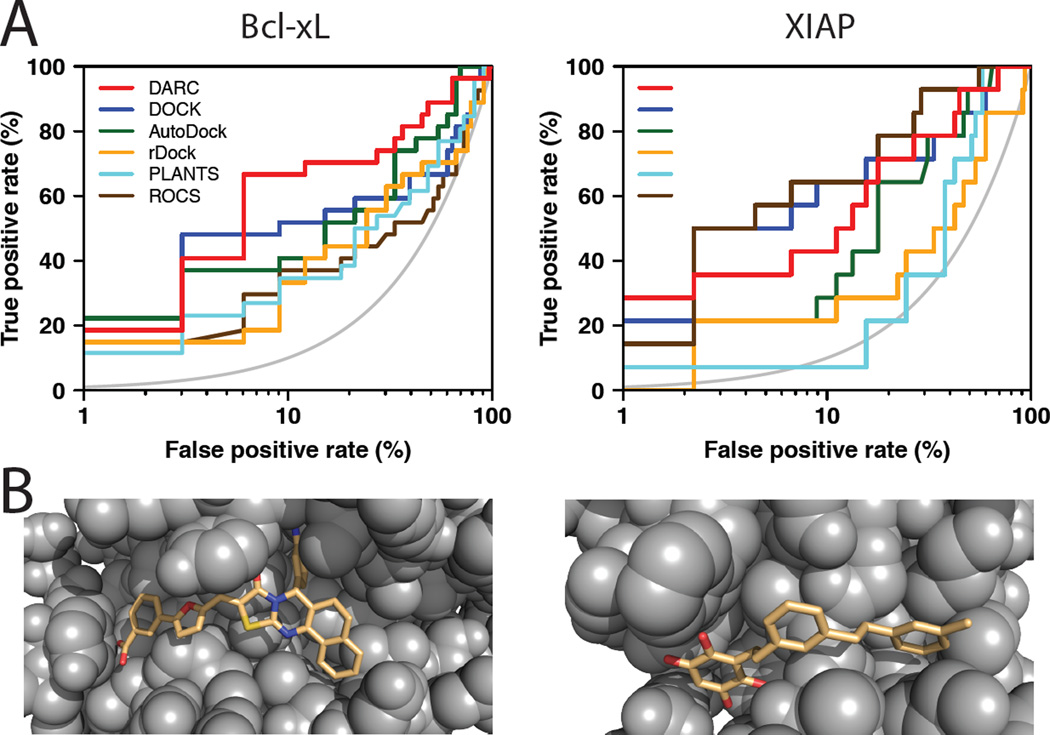
Figure 2. Virtual screening benchmark experiment using an inhibitor-bound protein structure.
(A) This receiver operating characteristic (ROC) plot compares the performance of various methods (DARC, DOCK [29,30], AutoDock [31], rDock [32], PLANTS [33], and ROCS [34–36]) for predicting whether a given compound is active against a particular protein interaction site. Drug-like “decoy” compounds were drawn from the Astex diverse set [23], then further filtered to remove any compounds that are similar in chemical structure to an active compound or any other decoy compound. The results are presented on a semi-log plot to highlight the “early” performance of the methods; the grey curve indicates the random retrieval of compounds (i.e. a random predictor). Left: discriminating 27 diverse compounds active against Bcl-xL from among 33 matched decoy compounds. Right: discriminating 14 diverse compounds active against XIAP from among 45 matched decoy compounds. (B) DARC-docked models of representative active compounds against Bcl-xL (left) and XIAP (right). These compounds were not chemically similar to those used in solving these crystal structures (such compounds were excluded from this benchmark).