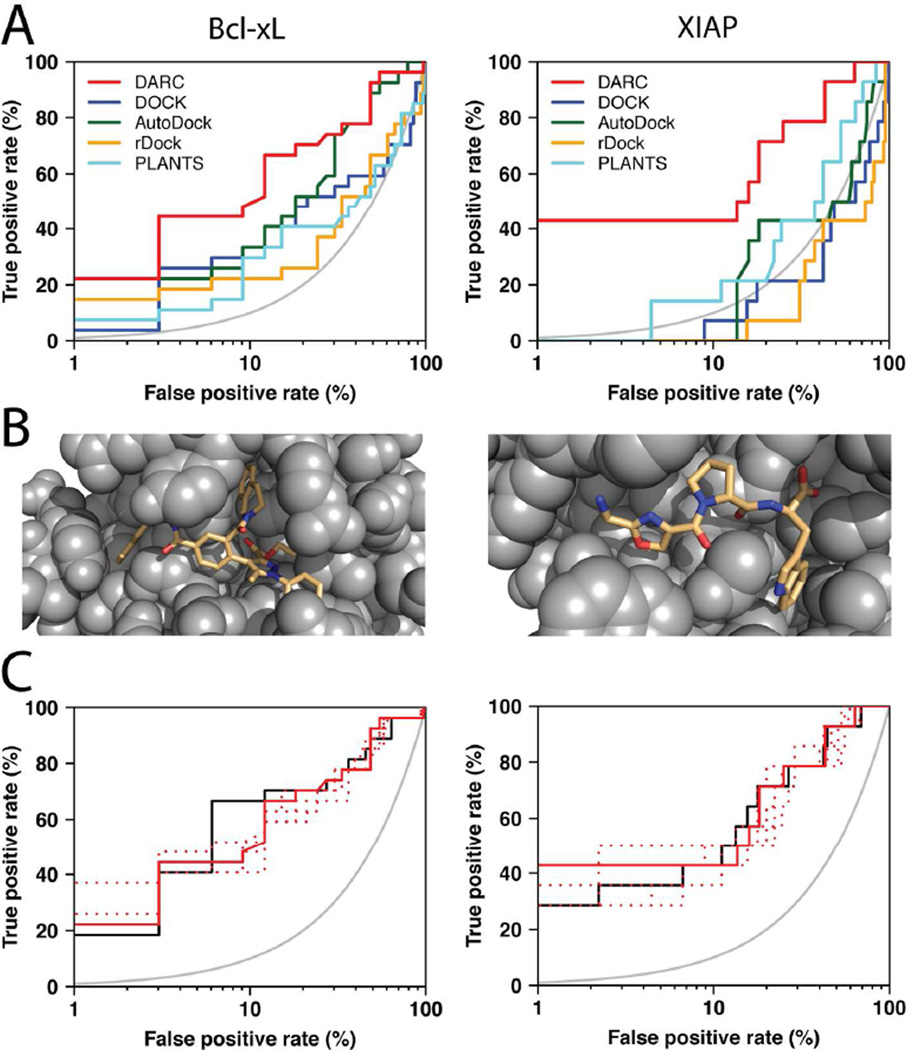
Figure 3. Virtual screening benchmark experiment using protein structures from “pocket optimization” simulations.
(A) This benchmark experiment involves discrimination of the same active versus “decoy” compounds as in Figure 2, however this time compounds were screened against a protein conformation generated via “pocket optimization” simulations initiated from an unbound crystal structure (instead of protein conformations from an inhibitor-bound crystal structure) [16]. The results are again presented on a semi-log plot to highlight the “early” performance of the methods; the grey curve indicates the random retrieval of compounds (i.e. a random predictor). (B) Models of representative active compounds docked by DARC to the “pocket optimized” conformation of Bcl-xL (left) or XIAP (right). (C) The performance of DARC in this benchmark is essentially equivalent when screening against a known ligand-bound structure (solid black line), the lowest-energy “pocket optimized” conformations (i.e. as described in the previous panels) (solid red line), or any of five other low-energy “pocket optimized” conformations (dashed red lines); this observation demonstrates the insensitivity of the method to details of the protein structure.