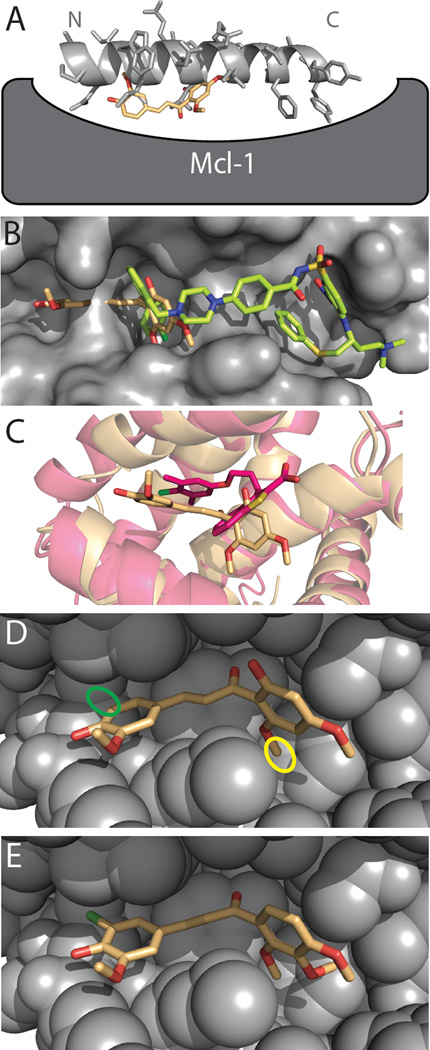
Figure 5. Evaluating DARC’s model of the M0/Mcl-1 complex.
(A) Though M0 was identified in a screen against a peptide-bound conformation of Mcl-1, M0 (peach) is distinctly not mimicking the interactions of the cognate peptide (light gray). In the DARC-generated model M0 competes with the N-terminus of the peptide for Mcl-1 binding, but fits more deeply into the Mcl-1 surface pocket than the peptide sidechains. (B) This M0 binding mode is distinctly different from that of known Bcl-xL inhibitors such as ABT-737 (green), which instead overlap with the C-terminus of the peptide-binding site. The surface of the Bcl-xL protein is shown (grey), solved in complex with ABT-737. (C) Other recently-described inhibitors of Mcl-1, including the representative example shown here (dark pink) [59], bind at a similar location to M0; however, each distinct series of inhibitors induce their own unique Mcl-1 conformational change to allow deeper ligand burial. (D) The locations of two M0 positions that play key roles in our model of the complex: the 6’-methoxy group (yellow) and the unsubstituted 5-position (green). (E) A model of the D1, the most potent M0 derivative, in complex with Mcl-1. This compound preserves the crucial 6’-methoxy group, and embellishes the 5-position with a chlorine substitution to improve packing. Closely analogous compounds to D1 that lack the methoxy group at the 6’-position (D3) or that lack the chlorine at the 5-position (D5) exhibit reduced activity.