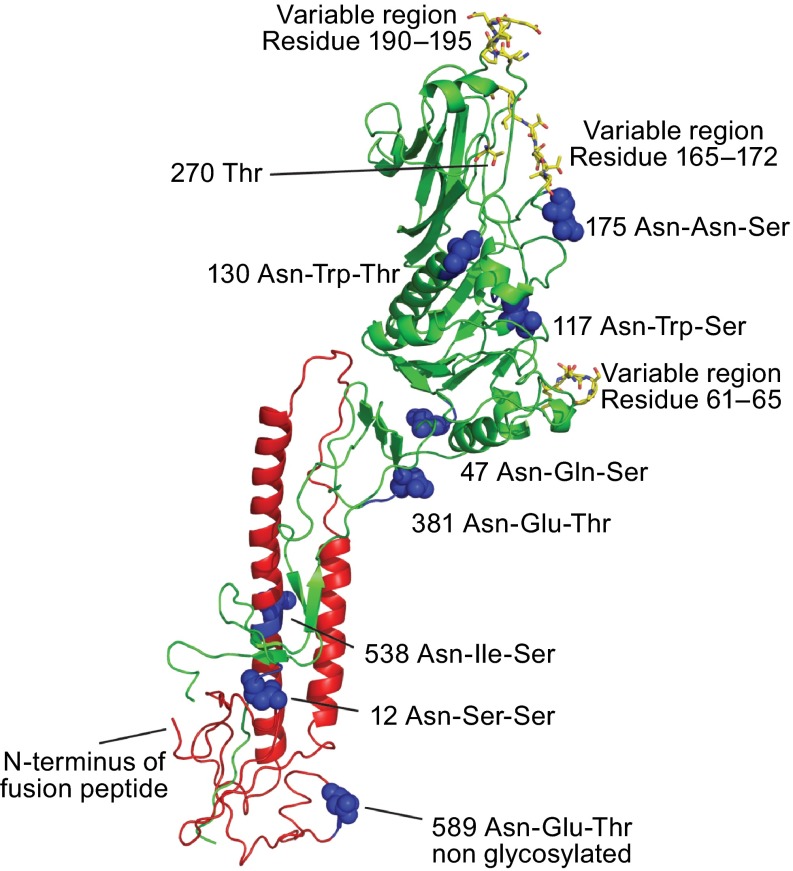
Figure 4.
Location of N-Glycosylation sites and variable regions in the crystal structure of HEF. Asparagine residues of used and unused glycosylation sequons (Asn-X-Ser/Thr) are depicted in the secondary structure of a HEF monomer as highlighted as balls. Amino acids in the HEF protein from C/JHB/1/66, which vary in other influenza C virus isolates are marked as sticks (Table S1). Threonine residue 270, which is exchanged by isoleucine in a virus variant that has acquired the ability to grow in MDCKII cells, is also marked as sticks. (This residues is termed Thr284 in the publication (Szepanski et al., 1992) since the 14 amino acid long signal peptide was included in the numbering). Figure was created with PyMol from PDB file 1FLC. HEF1 and HEF2 subunits are drawn in red and green, respectively