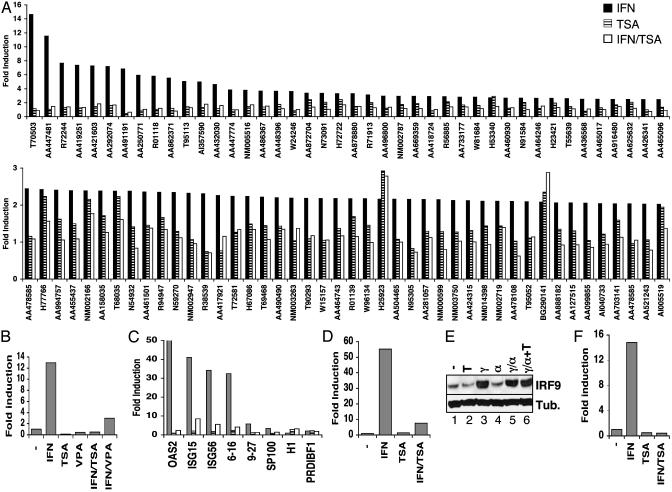
Fig. 1.
Global impairment of IFN-stimulated gene expression by HDAC inhibition. (A) U2OS cells were stimulated with IFNα in the absence (filled bars) or presence of TSA (open bars) or were treated with TSA alone (hatched bars) for 8 h. Isolated RNA was analyzed by two-color microarray hybridization in comparison with RNA isolated from untreated cells. A Upper represents genes induced by IFNα >2.5-fold; A Lower includes genes induced >2-fold. (B) HeLa cells were treated with IFNα, TSA, and VPA, as indicated, for 6 h before isolation of RNA. Expression of ISG54 was quantified by real-time RT-PCR and represented as fold induction over untreated cells. (C) Expression of additional ISG transcripts was analyzed as in B, except that HeLa cells were treated overnight with IFNγ; cells were treated with IFNα (filled bars), TSA (hatched bars), or IFNα plus TSA (open bars) for 4.5 h before isolation of RNA and analysis by quantitative RT-PCR with primers specific for the indicated genes. (D) As in B, except human fibroblast FS2 cells were treated for 1 h with IFNα and/or TSA, as indicated, before quantitation of ISG54 transcript levels. (E) Western blot analysis of IRF9 and β-tubulin expression in HeLa cells treated with TSA (T), IFNγ (γ), and IFNα (α), as indicated. (F) As in C, except expression of ISG54 nuclear RNA precursors rather than mature mRNA was quantified by using an intron/exon primer pair.