Abstract
Oxidative stress might contribute to the occurrence of cancers, including the hematological ones. Various genetic polymorphisms were shown to increase the quantity of reactive oxygen species, a phenomenon that is able to induce mutations and thus promote cancers. The purpose of the study was to evaluate the association between CAT C262T, GPX1 Pro198Leu, MnSOD Ala16Val, GSTM1, GSTT1, and GSTP1 Ile105Val gene polymorphisms and acute myeloid leukemia risk, in a case-control study comprising 102 patients and 303 controls. No association was observed between AML and variant genotypes of CAT, MnSOD, GSTM1, and GSTT1 polymorphisms. Our data revealed a statistically significant difference regarding the frequencies of GPX1 Pro198Leu and GSTP1 Ile105Val variant genotypes between AML patients and controls (p < 0.001). Our results showed no association in the distribution of any of the CAT C262T, GPX1 Pro198Leu, GSTM1, GSTT1, and GSTP1 polymorphisms regarding age, gender, FAB subtype, cytogenetic risk groups, FLT3 and DNMT3 gene mutations, and overall survival. Our data suggests that the presence of variant allele and genotype of GPX1 Pro198Leu and GSTP1 Ile105Val gene polymorphisms may modulate the risk of developing AML.
1. Introduction
Acute myeloid leukemia (AML) is a complex disease characterized by the accumulation of blasts in the bone marrow and uncontrolled proliferation, in which excess production of oxygen derived radicals compromises the antioxidant defense system thereby leading to oxidative stress [1]. Excessive cellular reactive oxygen species (ROS) or deficiencies in antioxidant defenses are generators of oxidative stress [2], and as previously demonstrated, tumor cells show higher susceptibility to oxidative stress, compared to normal cells.
Oxidative stress has been frequently observed in cancer and was also reported in several hematopoietic malignancies including acute lymphoblastic leukemia [3], acute myeloid leukemia [4], myelodysplastic syndrome (MDS), and chronic myeloid leukemia (CML) [5].
Elevated and persistent ROS levels produce oxidative DNA damage and lead to single-stranded and double-stranded DNA breaks, thus promoting mutagenesis [2].
According to Udensi and Tchounwou, ROS may lead to cancer development by causing gene mutations and/or chromosomal aberrations [6]. Increased production of ROS can lead to acquisition of genomic changes, thereby producing genomic instability. This environment can sustain tumor formation and disease progression [7, 8]. Sallmyr et al. suggested that FLT3/ITD mutations (FMS-like tyrosine kinase 3, internal tandem duplications) in acute myeloid leukemia (AML) result in ROS production [7]. This may lead to DNA damage and defective repair mechanisms in myeloid leukemia, besides additional chromosomal aberrations and gene mutations.
The antioxidant defense system includes MnSOD (manganese superoxide dismutase), GPX (glutathione peroxidase), and catalase (CAT) which inactivate ROS [1, 9].
Manganese superoxide dismutase (MnSOD), an antioxidant enzyme, has a critical role in protecting cells against oxidative stress by eliminating superoxide radicals after their conversion to H2O2 and oxygen [5, 10, 11].
Several MnSOD polymorphisms were found to cause an increased superoxide dismutase activity and increased H2O2 (hydrogen peroxide) quantities that may produce high levels of ROS, which if not subsequently neutralized represent contributing factors to the neoplastic transformation of cells [5, 12].
Glutathione peroxidase (GPX1), an antioxidant defense enzyme, detoxifies hydrogen peroxide into water and oxygen [13]. It has been reported that GPX1 gene polymorphism (GPX1 Pro198Leu) is associated with decreased enzyme activity, thereby conferring an increased risk of developing cancer in Caucasians [14].
Catalase (CAT), an important enzyme of the antioxidant system, converts H2O2 to water and molecular oxygen [15]. The expression of catalase is influenced by gene polymorphisms, which may lead to differences in susceptibility of individuals to oxidative damage caused by ROS [15]. According to an up-to-date meta-analysis performed by Shen et al., CAT C262T polymorphism may be a risk factor for cancer, with cancer type-specific effects [16].
Glutathione S-transferase (GST) enzymes are involved in the metabolism and detoxification of a wide variety of oxidative stress products, xenobiotics, and carcinogens [3].
Glutathione S-transferases (GSTs), phase II metabolizing enzymes, are encoded by GST genes and have an important role in cellular defense and therefore in protecting tissues against oxidative damage [5, 17].
GST gene polymorphisms are associated with deficiencies in enzyme activity and have been implicated in susceptibility to acute leukemia, but study results are still controversial [4, 18, 19].
To the best of our knowledge, even though the role of oxidative stress and GSTM1, GSTT1, GSTP1, CAT, and GPX1 as well as MnSOD2 in the pathogenesis of cancer have been previously investigated, no studies on the association of all these six polymorphisms with acute myeloid leukemia have been previously published.
The purpose of the study was to investigate possible associations between glutathione S-transferases (GSTM1, GSTT1, and GSTP1), superoxide dismutase (MnSOD Ala16Val), glutathione peroxidase (GPX1 Pro198Leu), catalase (CAT C262T) gene polymorphisms, and AML susceptibility in a Romanian population.
2. Material and Method
2.1. Patients and Controls
The research was conducted at the Department of Genetics, University of Medicine and Pharmacy of Targu Mures. The study group consisted of 102 unrelated AML patients (46 males and 56 females; mean age 51.70 ± 16.761 SD, standard deviation, years) and 303 (174 males and 129 females; mean age 46.46 ± 14.501 SD years) unrelated healthy controls with no known malignancies.
The patients were diagnosed with AML according to French-American-British (FAB) subtype and also according to WHO standards [20, 21] at the Hematology Clinics from Targu Mures, Romania, between 2010 and 2013. AML patients were stratified by French-American-British (FAB) subtype as follows: 9 M0, minimally differentiated AML (8.8%); 25 M1, AML without maturation (24.6%); 26 M2, AML with maturation (25.5%); 3 M3, acute promyelocytic leukemia (2.9%); 19 M4, acute myelomonocytic leukemia (18.6%); 15 M5, acute monocytic leukemia (14.7%); 1 M6, erythroleukemia (0.9%); and 4 M7, megakaryoblastic leukemia (3.9%).
Based on WHO 2008 standards and available data, AML cases included in the present study were classified as follows: 14 AML with recurrent cytogenetic anomalies (13.72%), 9 AML dysplasia related (8.82%), 0 myeloid neoplasia therapy related (0%), 0 myeloid sarcoma (0%), 0 myeloid proliferations Down syndrome related (0%), and 79 AML not otherwise specified (77.45%).
Regarding the cytogenetic risk group, favorable group comprised 10 AML patients (9.9%) and intermediate risk group comprised 66 cases (64.7%) while unfavorable (or high risk) cytogenetic group comprised 12 patients (11.7%).
For induction treatment, the standardized protocol consisting of a combination of anthracycline and cytarabine-ara (Ara-C) in the well-known 7 + 3 days' protocol was used. The anthracycline used was daunorubicin or idarubicin. Ara-C was administered as a bolus every 12 hours or continuous infusion over 7 days. All the transretinoic acid (ATRA) was administered in patients with a diagnosis of acute promyelocytic leukemia (APL). In AML, intensive consolidation was used, the standard dose for consolidation being 1.5 g/m2 every 12 hours. Reduced intensity conditioning was used in some cases in patients above 60 years of age or in those with poor performance (due to the associated diseases) before the start of chemotherapy.
Both patients and healthy controls were from the central region of Romania. AML cases were followed-up till their death or the beginning of 2015. From the investigated AML patients, 57 cases achieved complete remission (CR) from which 31 relapsed, 23 attained partial remission (PR), and 21 were refractory to treatment.
2.2. Genotyping Procedures
Fresh whole blood samples were collected at the time of diagnosis in tubes containing ethylene diamine triacetic acid (EDTA). Genomic DNA was extracted using Quick-gDNA MiniPrep kits (ZymoResearch, USA) and Wizard Genomic DNA Purification kits (Promega, Madison, WI, USA) according to the manufacturers' instructions.
FLT3 (fms-like tyrosine kinase 3) and DNMT3A (DNA methyltransferase) mutations were assessed using PCR-based methods in all AML patients, as previously described [22, 23].
CAT C262T, GPX1 Pro198Leu, MnSOD Ala16Val, and GSTP1 Ile105Val polymorphisms were genotyped by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) methods as previously described with minor modifications to the PCR protocol and digestion step for superoxide dismutase gene which consisted of exposing PCR amplicons obtained by Thermo Scientific FastDigest HaeIII (BsuRI) for 20 minutes [15, 24–26].
Genotyping of the GSTM1 and GSTT1 polymorphisms was carried out by multiplex polymerase chain reaction as described by Sharma et al. [27].
2.3. Statistical Analysis
Nominal variables were described as absolutes and relative frequencies (%) and the association between them was analyzed by Pearson's Chi-square test or Fisher's Exact Test. The size effect for statistically significant associations was expressed as an odds ratio (OR) with 95% confidence interval associated. Each polymorphism of interest was analyzed univariately as a possible predictor for AML using simple binary logistic regression. The independent effect of a polymorphism was tested using multiple binary logistic regression. Multivariable model was defined considering all exogenous variables whose estimated significance level in univariate logistic regression was p < 0.25, the polymorphism's effects being adjusted for possible confounders (gender variable). The best predictive model was chosen comparing nested models based on Akaike information criterion (AIC), Bayesian information criterion (BIC), and Likelihood Ratio Test (LR). All the regression models were additive models that resulted from no significance of interaction terms.
The performance of the final logistic model was evaluated with respect to goodness of fit (Nagelkerke R 2 coefficient, Brier score), discrimination (predictive and classificatory ability) [28], and calibration aspects [29]. Discrimination was established using c-index which is equal to the area under receiver operating characteristic curve (AUC), while Somers' D index, discrimination slope, and calibration were analyzed by graphical representation of the concordance between predicted model probabilities and observed proportions of criterion variable. The unreliability index (U), quality index (Q), maximal error, and mean squared error were used for measuring model miscalibration. In order to assess the reproducibility of the logistic model we performed internal validation by bootstrap resampling method [30]. Using 1000 bootstrap resamples, we calculated unbiased optimism corrected for all estimates of model performance indices.
The Kaplan-Meier estimates were determined for each polymorphism and survival distributions were compared using DeLong test.
The level of statistical significance for all two-sided tests was set at p < 0.05.
Statistical analysis was performed with the R software version 3.1.3 (R Foundation for Statistical Computing, Vienna, Austria) using rms libraries.
The study protocol was approved by the Ethics Committees of the University of Medicine and Pharmacy Tirgu Mures and informed consents were signed by the patients.
3. Results
FLT3 mutations were found in 24 AML cases while DNMT3A mutations were identified in 12 patients. The frequency of CAT 262T allele in AML cases was 25.9% while in controls it was 26.5% (p = 9.269). The frequency of GPX1 198Leu allele was significantly higher in AML cases (83.3%) compared to healthy controls (57.2%) (p < 0.0001, OR = 3.707, and 95% CI: 2.48–5.541). Variant allele frequencies of the MnSOD Ala16Val polymorphism in case and control groups were 47.3% and 54.5%. The frequency of variant GSTP1 105Val allele was 33.8% in AML group and 26.9% in control group. There was a significant difference between distributions of GSTP1 Ile105Val allele frequencies in AML group and control subjects (p < 0.0001, OR = 2.357, and 95% CI: 1.649–3.368). In AML patients, the frequencies of GSTM1 and GSTT1 null genotypes were 57.8% and 23.5%, respectively. The genotype distributions of CAT, GPX, MnSOD, GSTP1, GSTM1, and GSTT1 gene polymorphisms in AML cases and controls are shown in Table 1.
Table 1.
Distribution of CAT, GPX, MnSOD, GSTP1, GSTM1, and GSTT1 genotypes among AML patients and controls.
| Polymorphism | AML patients n (%) |
Controls n (%) |
p value, OR (95% CI) AML versus controls |
|---|---|---|---|
| CAT C262T | |||
| CC | 55 (53.9) | 161 (53.1) | Reference |
| CT | 41 (40.2) | 123 (40.6) | 0.985, 0.98 (0.61–1.56) |
| TT | 6 (5.9) | 19 (6.3) | 0.985, 0.92 (0.35–2.43) |
| CT + TT | 47 (46.1) | 142 (46.9) | 0.89, 0.969 (0.618–1.519) |
| GPX1 Pro198Leu | |||
| Pro/Pro | 3 (2.9) | 34 (11.2) | Reference |
| Pro/Leu | 28 (27.5) | 190 (62.7) | 0.588, 1.670 (0.480–5.80) |
| Leu/Leu | 71 (69.6) | 79 (26.1) | <0.0001, 10.186 (2.997–34.622) |
| Pro/Leu + Leu/Leu | 99 (97.1) | 269 (88.8) | 0.012, 4.171 (1.252–13.886) |
| MnSOD Ala16Val | |||
| Ala/Ala | 24 (23.5) | 54 (17.8) | Reference |
| Ala/Val | 60 (58.8) | 168 (55.4) | 0.464, 0.803 (0.457–1.413) |
| Val/Val | 18 (17.6) | 81 (26.7) | 0.074, 0.50 (0.248–1.009) |
| Ala/Val + Val/Val | 78 (76.5) | 249 (82.2) | 0.206, 0.705 (0.409–1.214) |
| GSTP1 Ile105Val | |||
| Ile/Ile | 39 (38.2) | 205 (67.7) | Reference |
| Ile/Val | 57 (55.9) | 88 (29.0) | <0.001, 3.405 (2.111–5.491) |
| Val/Val | 6 (5.9) | 10 (3.3) | 0.0393, 3.154 (1.083–9.183) |
| Ile/Val + Val/Val | 63 (61.8) | 98 (32.3) | <0.0001, 3.379 (2.120–5.386) |
| GSTM1 | |||
| Present | 43 (42.2) | 125 (41.3) | Reference |
| Null | 59 (57.8) | 178 (58.7) | 0.873, 0.964 (0.612–1.518) |
| GSTT1 | |||
| Present | 78 (76.5) | 240 (79.2) | Reference |
| Null | 24 (23.5) | 63 (20.8) | 0.56, 1.172 (0.686–2.002) |
There were no significant differences between variant genotype and AML risk for CAT C262T (Chi-square test, χ 2 = 0.03, df = 2, and p = 0.985), MnSOD2 (Chi-square test, χ 2 = 4.022, df = 2, and p = 0.134), GSTM1 (Chi-square test, χ 2 = 0.026, df = 1, and p = 0.873), and GSTT1 (Chi-square test, χ 2 = 0.339, df = 1, and p = 0.560) polymorphisms.
Our data revealed a statistically significant difference regarding the frequencies of GPX1 Pro198Leu genotypes between AML patients and controls (Chi-square test, χ 2 = 62.399, df = 2, and p < 0.001).
Also, a significant difference in the frequency of GSTP1 Ile105Val variant genotype was found between AML group and controls (Chi-square test, χ 2 = 27.606, df = 2, and p < 0.001).
The distribution of investigated polymorphisms' combined variant (heterozygous and homozygous) genotype in AML patients stratified by age, gender, FAB subtype, cytogenetic risk group, and FLT3 and DNMT3A mutations criteria is presented in Table 2.
Table 2.
AML patient characteristics according to the CAT C262T, GPX1 Pro198Leu, MnSOD Ala16Val, GSTP1 Ile105Val, GSTM1,and GSTT1 genotypes.
| CAT C262T | GPX1 Pro198Leu | MnSOD Ala16Val | GSTP1 Ile105Val | GSTM1 | GSTT1 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT + TT | p value | Pro/Pro | Pro/Leu + Leu/Leu | p value | Ala/Ala |
Ala/Val + Val/Val | p value | Ile/Ile |
Ile/Val + Val/Val | p value | Present | null | p value | Present | null | p value | |
| Gender | ||||||||||||||||||
| Female | 22 | 24 | Ref | 0 | 46 | Ref | 13 | 33 | Ref | 23 | 23 | Ref | 18 | 28 | Ref | 36 | 10 | Ref |
| Male | 33 | 23 | 0.264 | 3 | 53 | 0.055 | 11 | 45 | 0.308 | 16 | 40 | 0.026 | 25 | 31 | 0.574 | 42 | 14 | 0.699 |
| Age | ||||||||||||||||||
| <60 years | 37 | 31 | Ref | 1 | 67 | Ref | 17 | 51 | Ref | 24 | 44 | Ref | 24 | 44 | Ref | 53 | 15 | Ref |
| >60 years | 18 | 16 | 0.888 | 2 | 32 | 0.257 | 7 | 27 | 0.805 | 15 | 19 | 0.387 | 19 | 15 | 0.057 | 25 | 9 | 0.805 |
| Age (median, range) | ||||||||||||||||||
| <50 years | 27 | 20 | Ref | 1 | 46 | Ref | 13 | 34 | Ref | 16 | 31 | Ref | 17 | 30 | Ref | 35 | 12 | Ref |
| >50 years | 28 | 27 | 0.554 | 2 | 53 | 1.00 | 11 | 44 | 0.483 | 23 | 32 | 0.540 | 26 | 29 | 0.316 | 43 | 12 | 0.815 |
| FAB classification | 0.698 | 0.565 | 0.014 | 0.333 | 0.149 | 0.461 | ||||||||||||
| M0 | 2 | 4 | 0 | 6 | 2 | 4 | 2 | 4 | 1 | 5 | 6 | 0 | ||||||
| M1 | 15 | 10 | 2 | 23 | 2 | 23 | 10 | 15 | 10 | 15 | 18 | 7 | ||||||
| M2 | 16 | 10 | 0 | 26 | 10 | 16 | 5 | 21 | 12 | 14 | 19 | 7 | ||||||
| M3 | 2 | 1 | 0 | 3 | 1 | 2 | 1 | 2 | 1 | 2 | 3 | 0 | ||||||
| M4 | 11 | 8 | 0 | 19 | 6 | 13 | 11 | 8 | 5 | 14 | 16 | 3 | ||||||
| M5 | 6 | 9 | 1 | 14 | 0 | 15 | 6 | 9 | 11 | 4 | 10 | 5 | ||||||
| M6 | 1 | 3 | 0 | 4 | 1 | 3 | 2 | 2 | 1 | 3 | 3 | 1 | ||||||
| M7 | 2 | 2 | 0 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 1 | ||||||
| Cytogenetic risk group | 0.058 | 0.549 | 0.067 | 0.925 | 0.630 | 0.194 | ||||||||||||
| Favorable | 6 | 6 | Ref | 0 | 12 | Ref | 4 | 8 | Ref | 4 | 8 | Ref | 6 | 6 | Ref | 11 | 1 | Ref |
| Intermediate | 38 | 26 | 0.751 | 2 | 62 | 1.00 | 11 | 53 | 0.238 | 25 | 39 | 1.00 | 27 | 37 | 0.753 | 48 | 16 | 0.279 |
| Unfavorable | 2 | 8 | 0.204 | 0 | 10 | — | 5 | 5 | 0.665 | 4 | 6 | 1.00 | 3 | 7 | 0.730 | 6 | 4 | 0.136 |
| ND | 9 | 7 | 1.00 | 1 | 15 | 1.00 | 4 | 12 | 0.691 | 6 | 10 | 1.00 | 7 | 9 | 1.00 | 13 | 3 | 0.613 |
| FLT3 mutations | ||||||||||||||||||
| FLT3− | 40 | 38 | Ref | 3 | 75 | Ref | 17 | 61 | Ref | 31 | 47 | Ref | 36 | 42 | Ref | 57 | 21 | Ref |
| FLT3+ | 15 | 9 | 0.93 | 0 | 24 | 1.00 | 7 | 17 | 0.457 | 8 | 16 | 0.572 | 7 | 17 | 0.141 | 21 | 3 | 0.145 |
| DNMT3A mutations | ||||||||||||||||||
| DNMT3A− | 48 | 42 | Ref | 2 | 88 | Ref | 21 | 69 | Ref | 31 | 59 | Ref | 39 | 51 | Ref | 69 | 21 | Ref |
| DNMT3A+ | 7 | 5 | 0.744 | 1 | 11 | 0.316 | 3 | 9 | 1.00 | 8 | 4 | 0.054 | 4 | 8 | 0.510 | 9 | 3 | 1.00 |
ND: not determined; Ref: reference.
Presence of the variant genotype of CAT C262T, GPX1 Pro198Leu, MnSOD Ala16Val, and GSTP1 Ile105Val, as well as null GSTM1 and GSTT1 genotype in AML group was further analyzed in relation to gender and age. There was no significant difference regarding the variant genotype according to gender and age in AML patients (p > 0.05 for all these comparisons).
Furthermore, genotype frequencies for all six gene polymorphisms were compared with French-American-British (FAB) subtype and cytogenetic risk group.
No association was found between FAB subtype and some of the investigated gene polymorphisms, namely, CAT C262T, GPX1 Pro198Leu, GSTP1 Ile105Val, GSTM1, and GSTT1. A positive association was obtained between MnSOD Ala16Val variant heterozygous and homozygous genotype and FAB subtype (p = 0.014), precisely acute monocytic leukemia (M5).
In the case of cytogenetic classification according to the risk group, it was not associated with variant genotype of the investigated gene polymorphism (p > 0.5 for all comparisons performed).
Furthermore, we analyzed whether there are any associations between DNMT3A and FLT3 gene mutations and investigated gene polymorphisms in AML patients. We observed no significant differences (p > 0.05) between AML patients with DNMT3A or FLT3 gene mutations and variant genotype in the case of CAT C262T, GPX1 Pro198Leu, MnSOD Ala16Val, GSTP1 Ile105Val, GSTM1, and GSTT1.
We also investigated if there are any associations between AML patients' outcome and different parameters such as DNMT3A, FLT3 gene mutations, and variant genotypes of all investigated polymorphisms. Only FLT3 gene mutations were associated with patient outcome (Pearson Chi-Square test, p = 0.011). In addition, from multinomial logistic regression, the presence of FLT3 mutation was a predictor only for relapse (p = 0.005, OR = 4.95, 95% CI: 1.63–15.06, and reference category = CR) not for PR (p = 0.451, OR = 1.60, 95% CI: 0.47–5.45, and reference category = CR).
In addition, we analyzed the frequency of combined variant genotypes of CAT C262T, GPX1 Pro198Leu, MnSOD Ala16Val, GSTP1 Ile105Val, GSTM1, and GSTT1 in AML cases and controls.
Taking into account the observed frequencies of different combined genotypes of the investigated polymorphisms in relation to the number of variant genotypes, we tested the hypothesis of leukemia association with the presence of more than two variant genotypes. Our data revealed an association between the presence of leukemia and the presence of more than two variant gene polymorphisms (Chi-square test, χ 2 = 12.16, df = 4, and p = 0.016).
In case of the presence of 3 variant genotypes in our AML patients, the crude OR was 2.45 (95% CI crude OR: 1.23–4.88), in case of 4 variant genotypes the crude OR was 1.79 (95% CI crude OR: 0.89–3.60), in case of 5 variant genotypes the crude OR was 2.72 (95% CI crude OR: 1.20–6.19), and in case of 6 variant genotypes the crude OR was 5.79 (95% CI crude OR: 1.63–20.52).
The level of statistical significance of the estimated regression coefficient associated with each of the independent variables was estimated by univariate logistic regression and is described in Table 3. A statistically significant dependency relation was observed between three variables and leukemia (p < 0.05).
Table 3.
The effect of predictors on outcome variable (results from univariate logistic regression).
| Variables | Statistics Z | p ∗ | Crude OR | 95% CI for crude OR |
|---|---|---|---|---|
| Gender | 2,152 | 0,031 | 1,64 | 1,05–2,58 |
| CAT262 | −0,138 | 0,89 | 0,97 | 0,62–1,52 |
| MnSOD2 | −1,260 | 0,208 | 0,70 | 0,41–1.21 |
| GPX198 | 2,328 | 0,02 | 4,17 | 1,25–13,88 |
| GSTM1 | −0,160 | 0,873 | 0,96 | 0,61–1,52 |
| GSTT1 | 0,582 | 0,561 | 1,17 | 0,69–2,00 |
| GSTP1 | 5,118 | <0,001 | 3,38 | 2,12–5,39 |
∗Wald's test crude p values; reference categories: gender = women; CAT262 = normal; SOD2 = normal; GPX198 = normal; GSTM1 = present; GSTT1 = present; GSTP1 = present.
Table 4 presents the final logistic model, considered the best predictor of leukemia, consistent with the concerning data. From the set of all considered predictors, only GSTP1 Ile105Val polymorphism can be considered an independent predictor or an independent risk factor for leukemia (p < 0.0001) while GPX1 Pro198Leu and patient gender had a positive effect on the risk of leukemia but only with a tendency towards statistical significance (p = 0.09; p = 0.08, resp.).
Table 4.
The final multivariate logistic model.
| Variables | b ∗ | SE | p + | Adjusted OR | 95% CI for adjusted OR |
|---|---|---|---|---|---|
| Gender | 0,41 | 0,24 | 0,092 | 1,5 | 0,94–2,41 |
| SOD2 | −0,38 | 0,29 | 0,196 | 0,69 | 0,39–1,22 |
| GPX198 | 1,09 | 0,63 | 0,081 | 2,98 | 0,88–10,15 |
| GSTP1 | 1,12 | 0,24 | <0,0001 | 3,08 | 1,92–4,94 |
| Constant | −2,54 | 0,67 | 0,0001 | 0,08 | 0,02–0,26 |
∗Estimated unstandardized regression coefficients; SE = standard error; +Wald's test adjusted p value.
Figure 1 points out the intensity of the relation between model predictors (gender, GPX198, and GSTP1) and presence of AML leukemia, the existence of such a relationship being highlighted by a significance level of 0.05 and 0.10.
Figure 1.
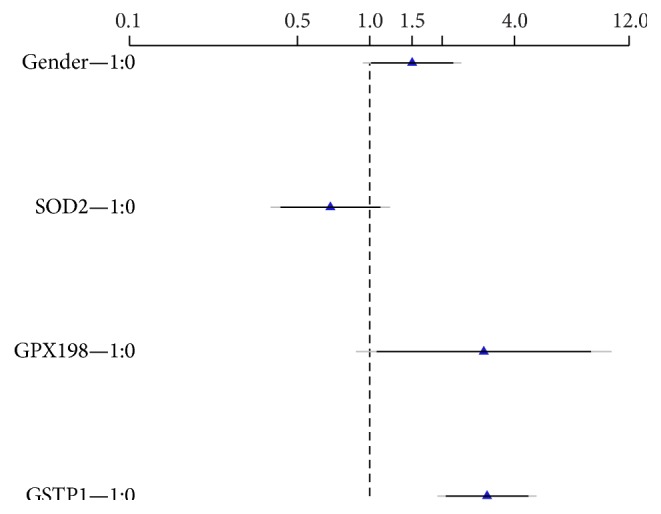
Graphical representation of estimated effects (OR) and associated confidence interval for each factor. Note: black = 90% confidence interval of multivariable adjusted OR; grey = 95% confidence interval of multivariable adjusted OR; 0 = reference category; 1 = variant category.
The model goodness-of-fit indices showed acceptable data fit, the discrimination indices revealed that the model had a good capacity to differentiate between AML subjects and healthy persons, the values close to zero of miscalibration indices showed a well-calibrated nomogram, and the internal validation procedure demonstrated the stability of selection of independent variables (Table 5).
Table 5.
Assessment of final model fit.
| Performance measure | Final predictive model | Internal validation∗ |
|---|---|---|
| Global measure of goodness of fit | ||
| Brier | 0,17 | 0,18 |
| R 2 (Nagelkerke) | 0,13 | 0,10 |
| Discrimination | ||
| AUC = C stat (95% CI) | 0,68 (0,63–0,75) | 0,67 |
| Somers' D index | 0,36 | 0,34 |
| Discrimination slope | 1,00 | 0,90 |
| Calibration | ||
| Hosmer-Lemeshow goodness-of-fit test (χ 2, p value) | 6,29 (0,39) | |
| Unreliability index U | −0,005 | 0,004 |
| Quality index Q | 0,09 | 0,06 |
| Maximal error | <0,001 | 0,04 |
| Mean squared error | 0,00061 | 0,00057 |
∗Evaluated by bootstrapping method (number of resampling, B = 1000); the optimism corrected indices values.
In Figure 2, the calibration graph suggested a predictive model with an acceptable level of concordance between predicted and observed probabilities.
Figure 2.
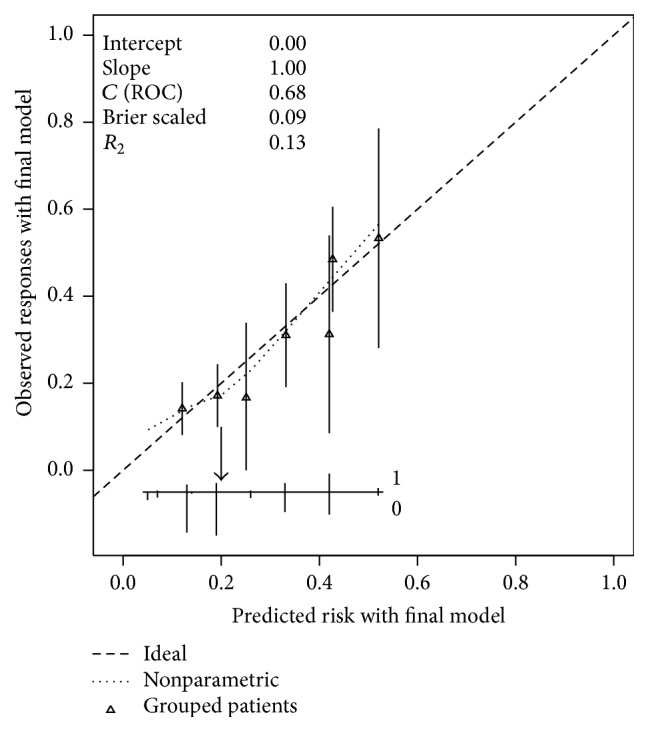
Calibration of the nomogram for AML prediction. The horizontal axis contains the predicted probability of AML; the vertical axis described the observed probability of AML. Perfect prediction corresponds to the dashed oblique line (45°). Bars correspond to CI of estimated grouped proportions and the arrow corresponds to the cut-off of 20% risk of AML.
4. Discussion
Certain genetic disorders, environmental carcinogens, physical (ionizing radiation) and chemical exposure, and chemotherapy may lead to acute myeloid leukemia, a heterogeneous disease [18, 31]. Sustained environmental stress may lead to overproduction of ROS and thus significant cell damage and occurrence of somatic mutations. This in turn favors the neoplastic transformation. MnSOD2, GPX, and CAT enzymes are involved in the prevention of DNA damage by ROS [32]. It was reported that MnSOD2, GPX, and CAT gene polymorphisms decrease the enzymatic activity and therefore may increase the risk of cancer by inducing oxidative DNA damage [14, 33].
Also, GST gene polymorphisms were associated with a decreased capacity of detoxification for certain mutagens and carcinogens [34].
In the present study, we investigated the oxidative stress enzyme polymorphisms on AML risk in a Romanian population, from the country's central region.
GST gene polymorphisms have been extensively studied in AML and GSTM1 and GSTT1 null genotypes have been found to increase the risk of AML in both Caucasians and Asians [4, 17, 18, 35, 36].
In the current study, we observed no significant differences between distributions of GSTM1 and GSTT1 null genotypes in patients with AML and controls. Similar results were found when we analyzed combined GSTM1 and GSTT1 null genotypes. Therefore, we may consider that GSTT1 and GSTM1 null genotypes are not associated with the risk of AML in Romanian patients. Our findings are similar to those reported in a recent study performed on chronic myeloid leukemia (CML) in a Romanian population [5].
In contradiction, a recent meta-analysis performed by He et al. showed that GSTM1 null genotype was associated with the risk of developing AML in East Asians while GSTT1 null genotype was a risk factor for AML in Caucasians [37]. The same study revealed that the presence of both GSTM1 and GSTT1 null genotypes might increase significantly the risk of AML in both Asians and Caucasians [37].
In a study performed on 147 ALL and 143 AML patients by Dunna et al., the homozygous variant genotype of the GSTP1 Ile105Val polymorphism was associated with the risk of developing acute leukemia and was associated with poor prognosis [19]. In agreement with the previous study, our research indicated that the presence of variant GSTP1 Ile105Val genotype significantly increases the risk of AML. In contradiction, in a meta-analysis performed by Tang et al. GSTP1 polymorphism was not associated with acute leukemia risk in Asians [18]. Similar results to those reported by Tang et al. were observed by He et al. [37].
Taking into account that the relationship between all six gene polymorphisms and overall survival has not been previously investigated, we analyzed the effect of predictors (variant genotype) on survival time by using DeLong's test crude p values.
Our findings revealed that GSTP1 Ile105Val variant genotypes and GSTT1 and GSTM1 null genotypes did not modify overall survival in AML patients.
Data obtained from our study show that the presence of variant genotypes of CAT C262T and MnSOD Ala16Val gene polymorphisms is not associated with the risk of AML. Our findings are consistent with a previous study performed on patients with CML from Romania [5]. Similarly, a recent case-study performed on Persian (Caucasians) Muslims living in Shiraz (Iran), a heterogeneous population, and a meta-analysis reported no significant association between CAT C262T gene polymorphism and susceptibility to breast cancer [38].
Results from Kaplan-Meyer analysis showed no difference in overall survival between patients with variant and wild-type genotype for CAT C262T gene polymorphism or for GPX1 Pro198Leu gene polymorphism.
Similar results were reported in a research performed on 89 AML patients regarding overall survival between carriers of the variant and wild-type genotypes of CAT C262T and GPX1 Pro198Leu polymorphisms [11].
Regarding MnSOD Ala16Val gene polymorphism and overall survival, our results showed that variant genotypes were not associated with significantly shorter survival in AML, compared to the wild-type genotype. Of the studied variables, only FLT3 mutation has a significant effect on survival time (Cox regression, p < 0.001).
In contradiction to our findings, in the study conducted by Koistinen et al. a significant overall survival (p = 0.02) was observed for AML patients carrying Val allele of MnSOD Ala16Val polymorphism compared to cases with Ala/Ala genotype [11].
Regarding GPX1 Pro198Leu genotypes, we observed that the presence of variant genotype is associated with a statistically significant risk of AML. Our findings are in contradiction with those previously observed on CML patients from the same region (central part) of Romania [5].
According to Liu et al. in a meta-analysis which included 8.102 patients with breast cancer no statistically significant association was found between MnSOD gene polymorphism and risk for breast cancer, excepting the variant allele in premenopausal women [39].
Our study is the first to evaluate the association of all six genes' polymorphisms (CAT C262T, GPX1 Pro198Leu, MnSOD Ala16Val, GSTT1, GSTM1, and GSTP1 Ile105Val) with AML among de novo patients.
Univariate logistic regression revealed that combined variant (heterozygous + homozygous) genotype of CAT, GPX1, MnSOD, and GSTP1 gene polymorphisms is associated with an increased risk of developing AML (p = 0.003, OR = 12.68, and 95% CI: 2.36–68.01).
A positive association was observed between combined variant (heterozygous + homozygous) genotype of CAT, GPX1, MnSOD, and GSTP1 and null genotype for GSTM1 and AML risk (p = 0.017, OR = 7.34, and 95% CI: 1.43–37.67). We have found a statistically significant correlation between variant genotypes for GPX1 Pro198Leu, MnSOD Ala16Val, GSTP1 Ile105Val, and GSTM1 null genotype and AML in our study (p = 0.004, OR = 10.33, and 95% CI: 2.12–50.26). Based on our data, we may suggest that the presence of one of the three possible combined genotypes may represent a risk factor for AML.
We could not find a statistically significant association in the distribution of any of the six gene polymorphisms regarding gender, cytogenetic risk group, and FLT3 and DNMT3A gene mutations.
To our knowledge, the relationship between the investigated gene polymorphisms and FLT3 and DNMT3A gene mutations and AML FAB subtype has not been previously investigated.
In addition, this study is the first on the distribution of CAT C262T, GPX1 Pro198Leu, MnSOD Ala16Val, GSTT1, GSTM1, and GSTP1 Ile105Val polymorphisms in Romanian AML patients.
Our study has some limitations, such as the relatively small group size, the lack of investigation of RUNX1 gene mutation (and other mutations), and the lack of antioxidant enzyme activity determination.
In conclusion, our present study reveals that the presence of variant allele and genotype of GPX1 Pro198Leu and GSTP1 Ile105Val gene polymorphisms may increase the risk of developing AML. As the number of patients is small, additional studies performed on larger cohorts are required to establish the relationship between these polymorphisms and AML risk.
Acknowledgment
This work was supported by Internal Research Grants of the University of Medicine and Pharmacy of Targu Mureş, Romania, Project no. 19/11.12.2013.
Conflict of Interests
The authors declare no conflict of interests.
Authors' Contribution
Claudia Bănescu designed the study, performed genetic analyses, and wrote the paper. Adrian P. Trifa contributed to the design of the study and performed critical revision of the paper. Erzsebet Benedek Lazar and Marcela Cândea collected blood samples and data for the AML patients. Carmen Duicu, Valeriu G. Moldovan, Florin Tripon, and Andrei Crauciuc performed genetic analysis and collected the data for controls. Mihaela Iancu performed statistical analysis. Minodora Dobreanu designed the research and performed critical revision of the paper. All authors read and approved the final version of the paper. Claudia Bănescu, Mihaela Iancu, Adrian P. Trifa, and Erzsebet Benedek Lazar equally contributed to this paper.
References
- 1.Naz A., Shamsi T. S., Sattar A., Mahboob T. Oxidative stress and total antioxidant status in acute leukemia at diagnosis and post remission induction phase. Pakistan Journal of Pharmaceutical Sciences. 2013;26(6):1123–1130. [PubMed] [Google Scholar]
- 2.Hole P. S., Darley R. L., Tonks A. Do reactive oxygen species play a role in myeloid leukemias? Blood. 2011;117(22):5816–5826. doi: 10.1182/blood-2011-01-326025. [DOI] [PubMed] [Google Scholar]
- 3.Moulik N. R., Parveen F., Kumar A., Agrawal S. Glutathione-S-transferase polymorphism and acute lymphoblastic leukemia (ALL) in north Indian children: a case-control study and meta-analysis. Journal of Human Genetics. 2014;59(9):529–535. doi: 10.1038/jhg.2014.66. [DOI] [PubMed] [Google Scholar]
- 4.Zhou L., Zhu Y.-Y., Zhang X.-D., Li Y., Liu Z.-G. Risk effects of GST gene polymorphisms in patients with acute myeloid leukemia: a prospective study. Asian Pacific Journal of Cancer Prevention. 2013;14(6):3861–3864. doi: 10.7314/apjcp.2013.14.6.3861. [DOI] [PubMed] [Google Scholar]
- 5.Bănescu C., Trifa A. P., Voidăzan S., et al. CAT, GPX1, MnSOD, GSTM1, GSTT1, and GSTP1 genetic polymorphisms in chronic myeloid leukemia: a case-control study. Oxidative Medicine and Cellular Longevity. 2014;2014:6. doi: 10.1155/2014/875861.875861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udensi U. K., Tchounwou P. B. Dual effect of oxidative stress on leukemia cancer induction and treatment. Journal of Experimental & Clinical Cancer Research. 2014;33(1, article 106) doi: 10.1186/s13046-014-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sallmyr A., Fan J., Rassool F. V. Genomic instability in myeloid malignancies: increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Letters. 2008;270(1):1–9. doi: 10.1016/j.canlet.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 8.Meitzler J. L., Antony S., Wu Y., et al. NADPH oxidases: a perspective on reactive oxygen species production in tumor biology. Antioxidants and Redox Signaling. 2014;20(17):2873–2889. doi: 10.1089/ars.2013.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasool M., Farooq S., Malik A., et al. Assessment of circulating biochemical markers and antioxidative status in acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) patients. Saudi Journal of Biological Sciences. 2015;22(1):106–111. doi: 10.1016/j.sjbs.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bag A., Bag N. Target sequence polymorphism of human manganese superoxide dismutase gene and its association with cancer risk: a review. Cancer Epidemiology Biomarkers and Prevention. 2008;17(12):3298–3305. doi: 10.1158/1055-9965.epi-08-0235. [DOI] [PubMed] [Google Scholar]
- 11.Koistinen P., Ruuska S., Säily M., et al. An association between manganese superoxide dismutase polymorphism and outcome of chemotherapy in acute myeloid leukemia. Haematologica. 2006;91(6):829–832. [PubMed] [Google Scholar]
- 12.Ji M., Tang J., Zhao J., Xu B., Qin J., Lu J. Polymorphisms in genes involved in drug detoxification and clinical outcomes of anthracycline-based neoadjuvant chemotherapy in Chinese Han breast cancer patients. Cancer Biology and Therapy. 2012;13(5):264–271. doi: 10.4161/cbt.13.5.18920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Alem U., Gann P. H., Dahl J., et al. Associations between functional polymorphisms in antioxidant defense genes and urinary oxidative stress biomarkers in healthy, premenopausal women. Genes and Nutrition. 2012;7(2):191–195. doi: 10.1007/s12263-011-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paz-y-Miño C., Muñoz M. J., López-Cortés A., et al. Frequency of polymorphisms pro198leu in GPX-1 gene and ile58thr in MnSOD gene in the altitude Ecuadorian population with bladder cancer. Oncology Research. 2010;18(8):395–400. doi: 10.3727/096504010x12644422320780. [DOI] [PubMed] [Google Scholar]
- 15.Suzen H. S., Gucyener E., Sakalli O., et al. CAT C-262T and GPX1 Pro198Leu polymorphisms in a Turkish population. Molecular Biology Reports. 2010;37(1):87–92. doi: 10.1007/s11033-009-9540-4. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y., Li D., Tian P., et al. The catalase C-262T gene polymorphism and cancer risk: a systematic review and meta-analysis. Medicine. 2015;94(13, article e679) doi: 10.1097/MD.0000000000000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He H.-R., Zhang X.-X., Sun J.-Y., et al. Glutathione S-transferase gene polymorphisms and susceptibility to chronic myeloid leukemia. Tumor Biology. 2014;35(6):6119–6125. doi: 10.1007/s13277-014-1810-7. [DOI] [PubMed] [Google Scholar]
- 18.Tang Z.-H., Zhang C., Cheng P., et al. Glutathione-S-transferase polymorphisms (GSTM1, GSTT1 and GSTP1) and acute leukemia risk in Asians: a meta-analysis. Asian Pacific Journal of Cancer Prevention. 2014;15(5):2075–2081. doi: 10.7314/apjcp.2014.15.5.2075. [DOI] [PubMed] [Google Scholar]
- 19.Dunna N. R., Vuree S., Kagita S., et al. Association of GSTP1 gene (I105V) polymorphism with acute leukaemia. Journal of Genetics. 2012;91(1):e60–e63. [PubMed] [Google Scholar]
- 20.Vardiman J. W., Thiele J., Arber D. A., et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 21.Tefferi A., Vardiman J. W. Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia. 2008;22(1):14–22. doi: 10.1038/sj.leu.2404955. [DOI] [PubMed] [Google Scholar]
- 22.Moreno I., Martín G., Bolufer P., et al. Incidence and prognostic value of FLT3 internal tandem duplication and D835 mutations in acute myeloid leukemia. Haematologica. 2003;88(1):19–24. [PubMed] [Google Scholar]
- 23.Brewin J. N., Horne G. A., Bisling K. E., Stewart H. J., Chevassut T. J. Rapid detection of DNMT3A R882 codon mutations allows early identification of poor risk patients with acute myeloid leukemia. Leukemia & Lymphoma. 2013;54(6):1336–1339. doi: 10.3109/10428194.2012.736986. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Zhang L., Sun D., Li Z., Wang L., Liu P. Genetic polymorphisms of superoxide dismutases, catalase, and glutathione peroxidase in age-related cataract. Molecular Vision. 2011;17:2325–2332. [PMC free article] [PubMed] [Google Scholar]
- 25.Manica-Cattani M. F., Cadoná F. C., de Oliveira R., et al. Impact of obesity and Ala16Val MnSOD polymorphism interaction on lipid, inflammatory and oxidative blood biomarkers. Open Journal of Genetics. 2012;2(4):202–209. doi: 10.4236/ojgen.2012.24026. [DOI] [Google Scholar]
- 26.Hohaus S., Di Ruscio A., Di Febo A., et al. Glutathione S-transferase P1 genotype and prognosis in Hodgkin's lymphoma. Clinical Cancer Research. 2005;11(6):2175–2179. doi: 10.1158/1078-0432.ccr-04-1250. [DOI] [PubMed] [Google Scholar]
- 27.Sharma A., Pandey A., Sardana S., Ashok S., Sharma J. K. Genetic polymorphisms of GSTM1 and GSTT1 genes in Delhi and comparison with other Indian and global populations. Asian Pacific Journal of Cancer Prevention. 2012;13(11):5647–5652. doi: 10.7314/apjcp.2012.13.11.5647. [DOI] [PubMed] [Google Scholar]
- 28.Steyerberg E. W., Vickers A. J., Cook N. R., et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21(1):128–138. doi: 10.1097/ede.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steyerberg E. W., Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. European Heart Journal. 2014;35(29):1925–1931. doi: 10.1093/eurheartj/ehu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steyerberg E. W., Harrell F. E., Jr., Borsboom G. J. J. M., Eijkemans M. J. C., Vergouwe Y., Habbema J. D. F. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. Journal of Clinical Epidemiology. 2001;54(8):774–781. doi: 10.1016/s0895-4356(01)00341-9. [DOI] [PubMed] [Google Scholar]
- 31.Coriu D., Jardan D., Jardan C., Tălmaci R., Dragomir M., Coliţă A. A new assay to identify recurrent mutations in acute myeloid leukemia using next-generation sequencing. Revista Română de Medicină de Laborator. 2014;22(1):93–99. doi: 10.2478/rrlm-2014-0003. [DOI] [Google Scholar]
- 32.Reuter S., Gupta S. C., Chaturvedi M. M., Aggarwal B. B. Oxidative stress, inflammation, and cancer: how are they linked? Free Radical Biology and Medicine. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuzhalin A. E., Kutikhin A. G. Inherited variations in the SOD and GPX gene families and cancer risk. Free Radical Research. 2012;46(5):581–599. doi: 10.3109/10715762.2012.658515. [DOI] [PubMed] [Google Scholar]
- 34.Karkucak M., Yakut T., Gulten T., Ali R. Investigation of GSTP1 (Ile105Val) gene polymorphism in chronic myeloid leukaemia patients. International Journal of Human Genetics. 2012;12(3):145–149. [Google Scholar]
- 35.Voso M. T., Fabiani E., D'Alo' F., et al. Increased risk of acute myeloid leukaemia due to polymorphisms in detoxification and DNA repair enzymes. Annals of Oncology. 2007;18(9):1523–1528. doi: 10.1093/annonc/mdm191. [DOI] [PubMed] [Google Scholar]
- 36.Seedhouse C., Faulkner R., Ashraf N., Das-Gupta E., Russell N. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clinical Cancer Research. 2004;10(8):2675–2680. doi: 10.1158/1078-0432.ccr-03-0372. [DOI] [PubMed] [Google Scholar]
- 37.He H.-R., You H.-S., Sun J.-Y., et al. Glutathione S-transferase gene polymorphisms and susceptibility to acute myeloid leukemia: meta-analyses. Japanese Journal of Clinical Oncology. 2014;44(11):1070–1081. doi: 10.1093/jjco/hyu121. [DOI] [PubMed] [Google Scholar]
- 38.Saadat M., Saadat S. Genetic polymorphism of CAT C-262 T and susceptibility to breast cancer, a case-control study and meta-analysis of the literatures. Pathology & Oncology Research. 2015;21(2):433–437. doi: 10.1007/s12253-014-9840-4. [DOI] [PubMed] [Google Scholar]
- 39.Liu G., Sun G., Wang Y., Wang D., Hu W., Zhang J. Association between manganese superoxide dismutase gene polymorphism and breast cancer risk: a meta-analysis of 17,842 subjects. Molecular Medicine Reports. 2012;6(4):797–804. doi: 10.3892/mmr.2012.998. [DOI] [PubMed] [Google Scholar]


