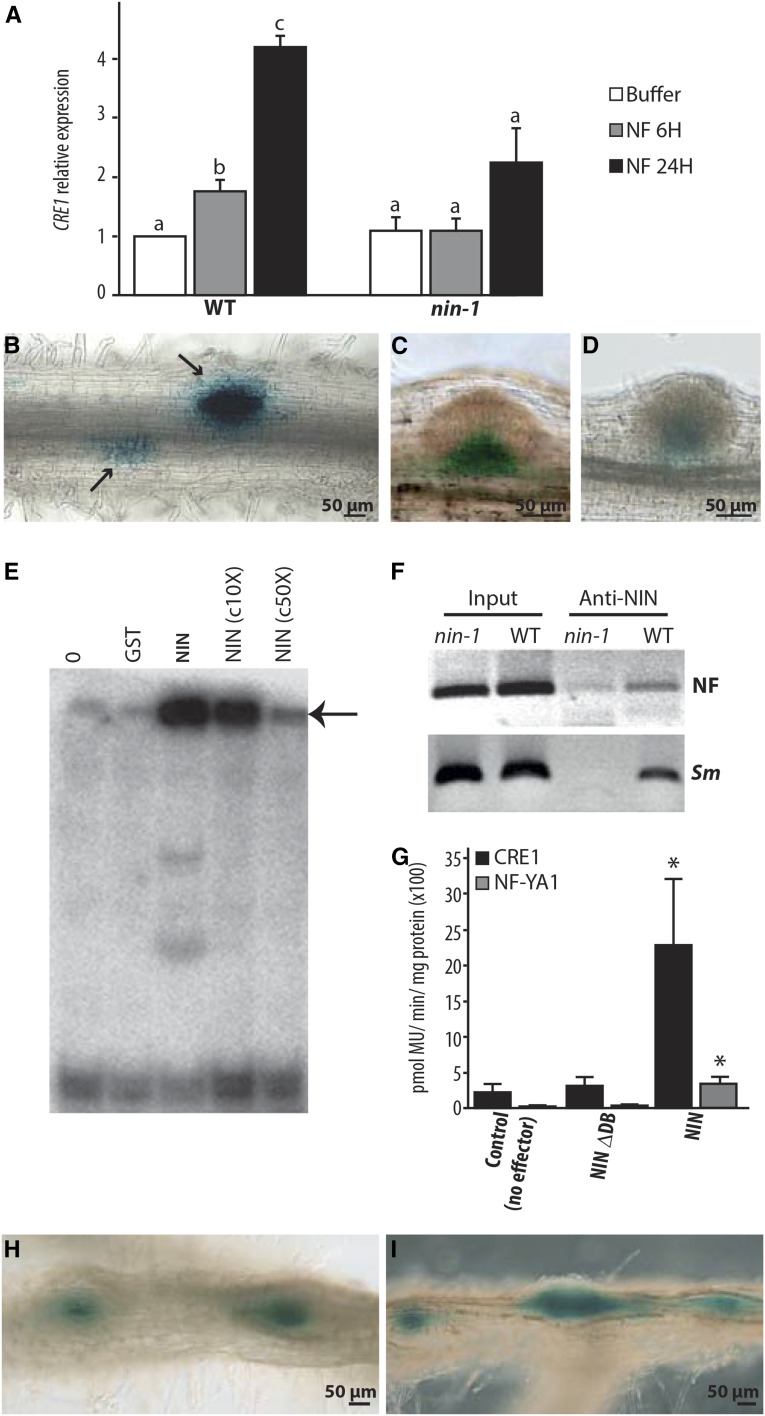
Figure 4.
NIN Directly Regulates CRE1 Expression.
(A) CRE1 induction by NF requires NIN as evidenced by RT-qPCR analysis. CRE1 expression was normalized with ACTIN11. The bars represent the ratio relative to the buffer control. Error bars indicate sd for three biological replicates. Different letters indicate statistical differences as determined by pairwise t tests in each genotype (P < 0.05).
(B) to (D) CRE1 expression measured using the CRE1 promoter driving the expression of GUS in the wild type ([B] and [C]) and in the nin-1 mutant (D), inoculated with S. meliloti. In wild-type roots, CRE1 is specifically expressed in response to rhizobia in cortical cells (arrows), while nonsymbiotic expression of CRE-1 is observed in the base of lateral root primordia (C). In nin-1 (D), only the nonsymbiotic expression of CRE1 is found in the base of lateral root primordia.
(E) NIN binds a radiolabeled CRE1 promoter probe (−1487 to −971), causing its retardation, indicated with an arrow. c10X and c50X, unlabeled competitor DNA in 10- and 50-fold excess reduce this degree of binding. Lane 0, no proteins incubated with radiolabeled probe; lane GST, GST protein incubated with radiolabeled probe.
(F) In vivo association of NIN with the CRE1 promoter measured using ChIP. Wild-type and nin-1 roots were treated with 1 nM NF for 24 h or with S. meliloti (Sm).
(G) Transactivation studies in N. benthamiana cells transiently transformed with the pCRE1:GUS or pNF-YA1:GUS reporters and respective effectors. The asterisk indicates a statistically significant difference (P < 0.05, Student’s t test) compared with the reference. NIN-∆DB, NIN carrying an internal deletion that removes the DNA binding domain.
(H) and (I) CRE1:GUS expression observed in wild-type roots expressing pEXPA:gNIN (H) or pNRT1.3:gNIN (I). GUS expression (in blue) is localized in spontaneous nodule-like structures.