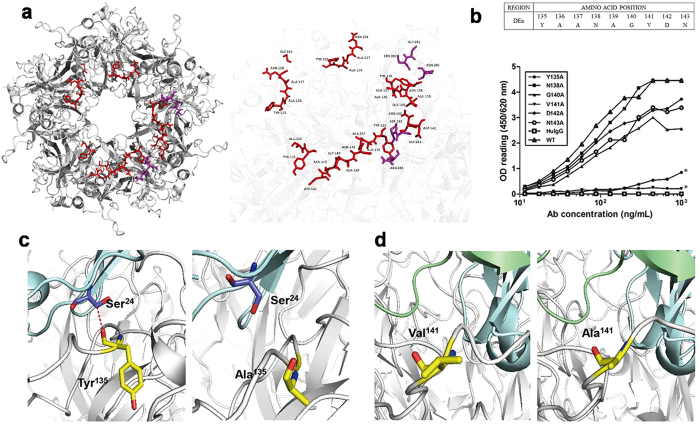
Figure 4. Identification of residues on the DEa loop that are critical for 26D1 binding.
(a) Localization of the epitopes for 26D1 binding predicted by the docking models. Predicted binding residues were predominantly located on the DEa loop with a few on the FGb loop. The antigen-antibody complex is displayed as a solid ribbon diagram (left). Binding residues in the DEa loop and the FGb loop are rendered in ball-and-stick, colored in red and purple, respectively (right). (b) Binding of 26D1 to DEa loop mutants of HPV16 L1 VLPs. Alanine scanning mutagenesis was conducted on the DEa loop except the parental Ala residues. Binding of 26D1 to the six point-mutated VLPs was detected in a binding assay. HPV16 wild-type VLPs were used as a positive control, and a human IgG1 antibody was used as a negative control. Asterisks indicate a significant difference in binding. (c) Tyr135 forming hydrogen bond with Ser24 of 26D1 (left); the Y135A substitution resulted in hydrogen bond disappearance and a substantially lower affinity to 26D1 (right). (d) The side chain of Val141 on the DEa loop pointed to the interior of the proteins (left). The V141A substitution resulted in a hydrophilic interface (right), Y135A and V141A also led to an energy elevation of 0.64 kcal/mol and 0.79 kcal/mol, which is consistent with the reduced binding affinity. Binding residues are rendered in ball-and-stick, and residues of 26D1 Fab and DEa loop are marked with blue and yellow, respectively. Models of 26D1 Fab and HPV16 antigen are displayed as solid ribbon diagrams. The red dashed line between residues (Tyr135 on DEa loop, Ser24 on 26D1 Fab) represents a hydrogen bond.